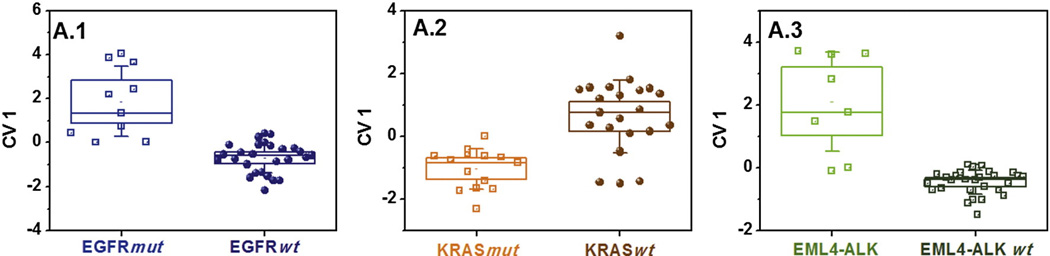
Figure 2.
Primary test set. DFA plots of CV1 that was calculated from the responses of the GNP sensors to the headspace samples of NSCLC cell-lines having EGFRmut, KRASmut or EML4-ALK oncogenes, or genetic mutations that are wt to the former three. The SD of the CV1 values is represented by the error bars. The boxes represent the 95% CI of the CV1 values, corresponding to 1.96*SE. The subpanels A.1-A.3 represent the three primary, general DFA models that are applied simultaneously to the data and contain all 37 headspace samples. Each point represents one sample. The samples were obtained from multiple cell-lines having the oncogenes of interest (cf. Table 1). Note that the CV1 distribution for the same group of samples may look different for two different DFA models due to different input parameters.