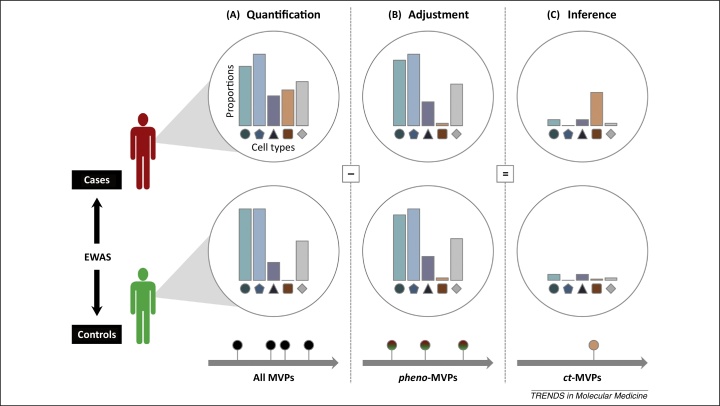
Figure 1.
The relevance of cellular heterogeneity in EWASs. Statistical methods that assess and adjust for the proportions of different cell populations derived from the DNA methylation profile of a cell have emerged as powerful tools in epigenome-wide association studies (EWASs). Some algorithms do not depend on reference data sets 5, 6, whereas other algorithms use reference data sets consisting of major leukocyte cell types [4]. Additional data sets will likely be integrated once released by the International Human Epigenome Consortium (http://ihec-epigenomes.org/). In general, the use of reference data sets is preferred because the cellular composition of a sample can be estimated more accurately. Below, we distinguish between three types of DNA methylation variable positions (MVPs), all of which are informative in different contexts. (A) Without correction of cellular heterogeneity, identified DNA methylation changes between cases and controls inform the overall heterogeneity of the phenotype of interest (irrespective of the underlying source and mechanism). (B) If adjustment for confounding cellular heterogeneity is performed, identified MVPs most accurately reflect methylation changes relevant to the phenotype. Such pheno-MVPs can subsequently be grouped into functionally more relevant differentially methylated regions (DMRs). (C) The contrast of all MVPs identified (A) with pheno-MVPs (B) may further characterise the disease state. For example, in addition to epigenetic changes that are related to the phenotype, an increase in lymphocyte populations may offer valuable clues to the immunobiology of the disease state [4]. Indeed, MVPs attributed to differential cell type distribution (termed ct-MVPs) may also shed light on the potential involvement of additional (or thus far disregarded) cell types in disease pathogenesis.