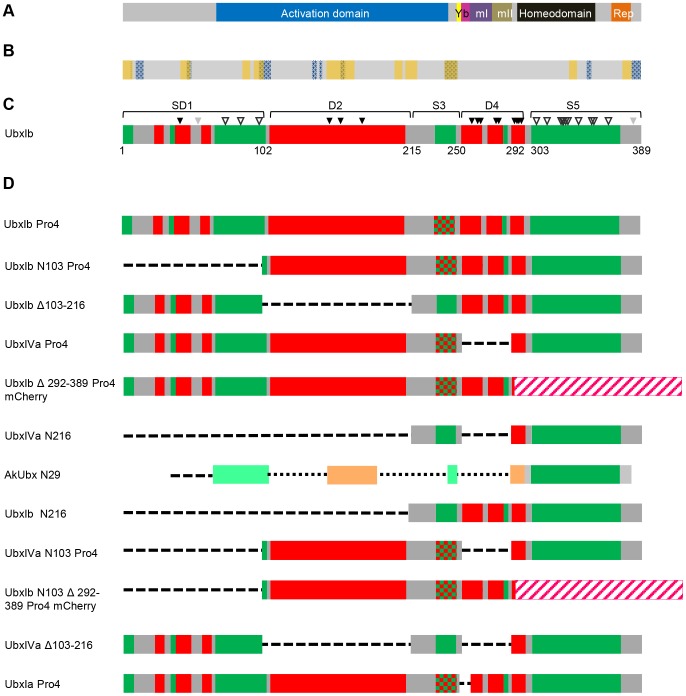
Figure 1. Location of structured and disordered regions in UbxIb, and design of Ubx variants.
(A) A grey bar, representing the domain organization of the UbxIb transcription factor shows the position of its transcription activation domain (blue), YPWM Exd interaction motif (yellow), DNA-binding homeodomain (black), a partial transcription repression domain (orange), and protein regions encoded by three alternatively spliced microexons: the b element (pink), mI (purple), and mII (brown). (B) The location of predicted protein-interaction motifs in Ubx as predicted by ANCHOR (yellow stripes) and MoRFpred (blue stippled stripes). Regions predicted by both algorithms to be involved in protein interactions are marked with both yellow and blue. (C) A bar schematic depicting the positions of structured and intrinsically disordered regions in UbxIb. The boundaries were determined by a combination of computational and experimental approaches. The scores from three disorder prediction algorithms were averaged to identify structured (green) and disordered (red) regions. Native state proteolysis, in which only disordered segments can be cleaved by trypsin, was used to verify these assignments, and, where appropriate, slightly expanded the boundaries of the predicted disordered regions [6]. Sites cut by trypsin (black triangles), sites not cut by trypsin (open triangles), and sites that could not be definitively assigned (grey triangles) are indicated. (D) Bar schematic for predicted protein interfaces and molecular recognition features (MoRFs) on Ubx peptide. The schematic bars show Anchor algorithm predicted Ubx- partner protein interfaces (orange bars) and MoRF algorithm predicted Ubx-partner protein interface (blue bars with pattern fill). (D) Bar schematics of Ubx truncation mutants and internal deletion mutants used in yeast two-hybrid assays to identify partner binding interfaces. UbxIb, UbxIa, and UbxIVa are isoforms created by alternative splicing in vivo. To prevent auto-activation, the activation domain was de-activated either by removal of amino acids 102 to 216 or by the Pro4 mutation, in which Ala and Glu are mutated to Pro at amino acids 226 and 233 (indicated by a red-green stipple), respectively, which should prevent formation of a predicted α-helix required for transcription activation [43]. In two variants, the structured C-terminus of the protein was replaced by mCherry, represented by a pink/white striped bar.