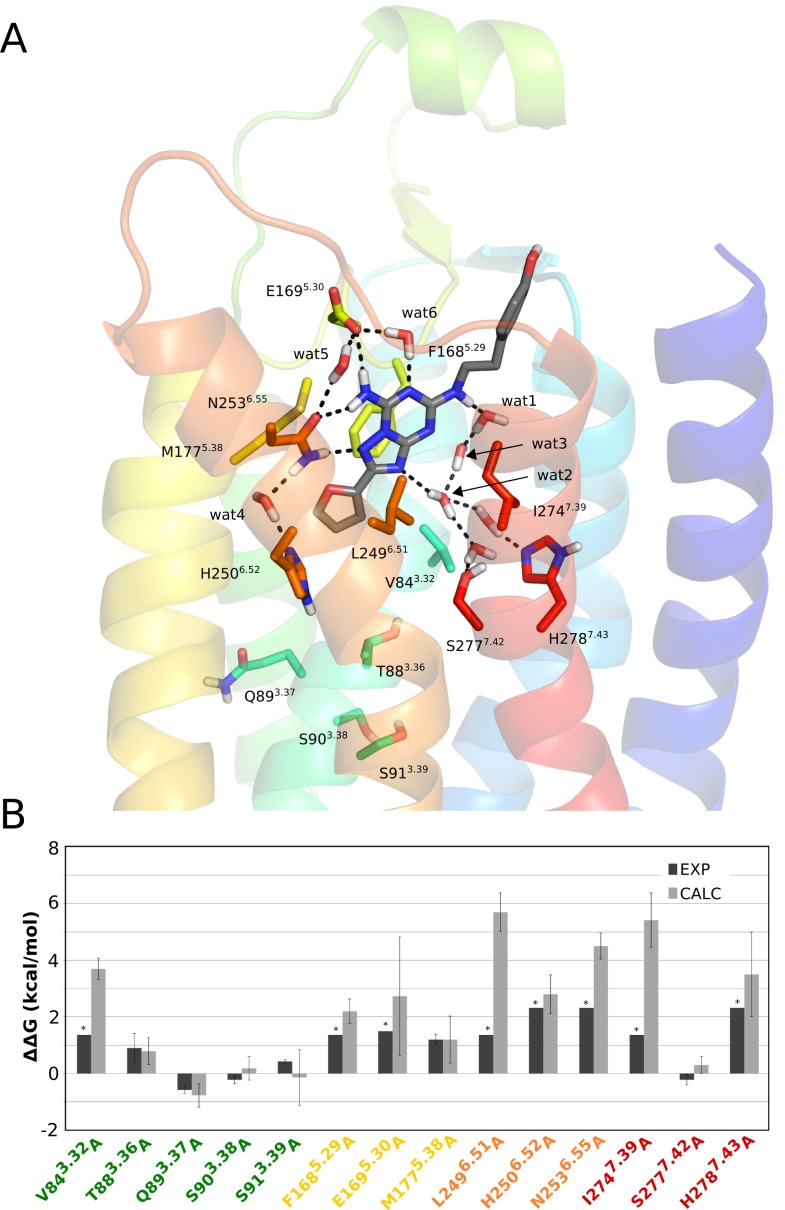
Figure 2. Structure of the A2AAR−ZM241385 complex and relative ligand binding free energies for mutants.
(A) Starting structure used for the FEP simulations with TM helices shown and colored according to a rainbow representation in anti-clockwise order (TM1 = blue → TM7 = red). Residues subjected alanine mutation are depicted in sticks, together with crystal water molecules mediating receptor-ligand interactions, and dashed lines indicate hydrogen bonds. (B) Calculated (gray bars) and experimental (black bars) relative binding free energies (kcal/mol) for ZM241385 to the fourteen A2AAR alanine mutants compared to the wt receptor. The star symbol denotes that an experimental value could not be determined and approximates the detection threshold of the experiment.