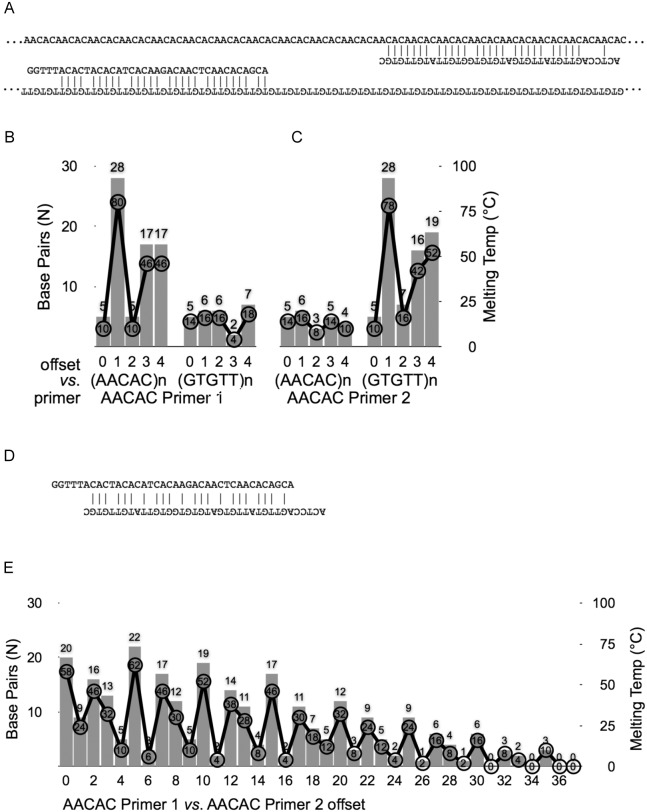
Figure 1. Computational validation of qPCR primers directed at AACAC satellite.
(A) Representation of homogenous block of AACAC repeat denatured and annealing to AACAC Forward and AACAC Reverse. Vertical lines indicate base pairs. (B) AACAC Forward hypothetically annealing to each of the five “phases”of AACAC (i.e., AACAC, ACACA, CACAA, ACAAC, and CAACA) and it’s reverse-complement. Bars indicate the number of base pairs possible in that phase, and line indicates ad hoc “melting” analysis. AACAC Forward has a strong preference for one phase and no affinity for the reverse-complement strand of the repeat. (C) as in (B), but with AACAC Reverse. (D) Shows the best possible pairing between AACAC Forward and Reverse. (E) AACAC Forward and AACAC Reverse pairing with every possible degree of overlap, from completely (at “0 offset”) to single 3′-most nucleotides pairing (at “37 offset”). Bars and lines are as in (B–C). In no case is AACAC Forward/Reverse dimer preferred over annealing to genomic targets (A–C) or the product of previous amplification.