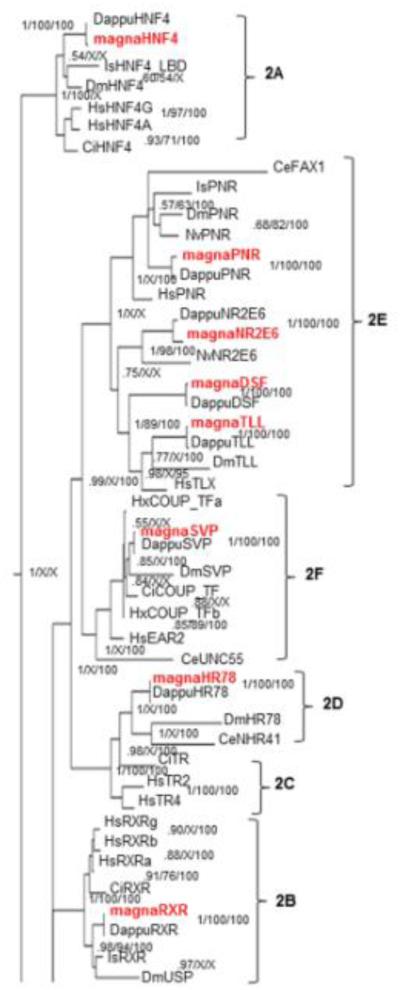
Fig 7. Phylogenetic relationship of nuclear receptors in subfamily 2.
The subfamily 2 nuclear receptors from different species were subjected to phylogenetic comparisons using Bayesian Inference, Maximum Parsimony, and Neighbor-Joining methods. The Bayesian tree is shown with posterior probabilities from the Bayesian tree, and bootstrap support values (frequency of occurrence) from the Neighbor-Joining and Maximum Parsimony trees provided in order from left to right, respectively. Probability values are separated by forward slashes at each corresponding node; an X indicates an area of disagreement from the Bayesian tree. Species included are D. pulex (Dappu), D. magna (magna), D. melanogaster (Dm), H. sapiens (Hs), I. scapularis (Is), C. elegans (Ce), C. intestinalis (Ci), and , N. vitripennis (Nv). Numbers at nodes are posterior probabilities. All D. magna sequences are in red.