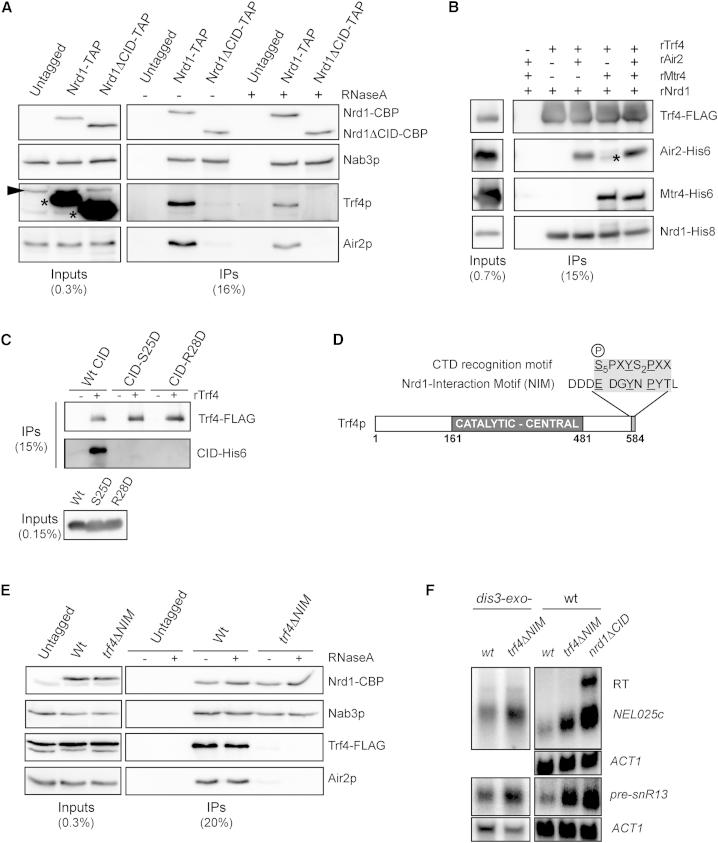
Figure 2.
Direct Interaction between Trf4p and Nrd1p Is Mediated by the CID
(A) Western blot analysis of Nrd1p-TAP and Nrd1ΔCIDp-TAP immunopurified complexes (IP). Samples were eluted by cleavage with the TEV protease. The indicated proteins were detected with specific antibodies, except for Nrd1p, which was detected with an anti-CBP antibody. The Trf4 signal in the input (indicated by an arrowhead) is partially overlapping with the Nrd1-TAP signal (denoted by an asterisk). The fraction of extract and immunoprecipitated material that is loaded on the gel is indicated.
(B) Immunoblot analysis of pull-down experiments performed with recombinant Trf4-FLAG as bait and E. coli extracts expressing recombinant His-tagged Air2, Mtr4, or Nrd1 as indicated. Immunoprecipitations were performed in the presence of RNase. An asterisk indicates a degradation fragment of Mtr4-His6.
(C) Immunoblot analysis as in (B) using recombinant Trf4-FLAG and recombinant CID-His6 or CID mutant derivatives (rCID-S25D-His6 and CID-R28D-His6), defective for the interaction with the CTD. Proteins were detected with an antibody anti-His tag or anti-FLAG.
(D) Scheme of Trf4p indicating the position and sequence of the NIM, compared to a CTD pattern containing the amino acids that mediate major contacts with the CID, including Ser5P (equivalent CTD and NIM regions are shaded; identical amino acids are underlined). Note the presence of a Ser5 phosphomimic (E) in the NIM.
(E) Western blot analysis of Nrd1-TAP immunopurified complexes from a TRF4 or trf4ΔNIM strain as in (A).
(F) Stabilization of NNS targets in trf4ΔNIM cells. Analysis of NEL025C and pre-snR13 RNAs by northern blot in the presence or absence of the NIM, in an exosome-defective (Dis3p catalytic mutant, dis3-exo−; left panels) or an otherwise wild-type background (right panels). Note that the rightmost panels were more exposed than the other panels to visualize the poorly detectable NEL025C and pre-snR13 transcripts in a strain wild-type for the nuclear exosome. Stabilization values in dis3-exo−/trf4ΔNIM relative to dis3-exo− are 1.9 ± 0.37 and 2.5 ± 0.56 for pre-snR13 and NEL025c, respectively (average and SD from three independent samples). Stabilization of pre-snR13 and NEL025c was consistently observed in trf4ΔNIM cells but could not be reliably quantified due to the low levels of these RNAs in a wild-type background.