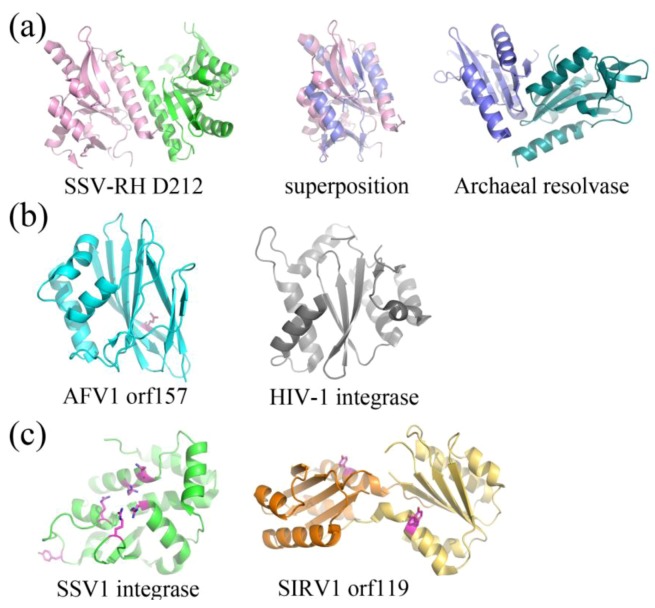
Figure 6.
Fold Topologies from archaeal viral enzymes involved in nucleic acid metabolism. (a) SSV-RH D212 (left, PDB ID 2W8M [35]) with a fold that is similar to those of members belonging to the PD-D/EXK nuclease superfamily. An archaeal member of this family, HJC Holliday junction resolving enzyme from Sulfolobus solfataricus (right, PDB ID 1HH1 [40]) aligns quite well with a single D212 subunit (middle), however the dimer interfaces are substantially different for the two. Each subunit within the dimer is colored differently. (b) AFV1 orf157 (left, PDB ID 3II2 [34]) very distantly resembles the two-layer sandwich fold of the phosphonucleotidyl superfamily to which HIV1-integrase belongs (right, PDB ID 1ITG [41]), however it only has one layer of α-helices and not two. A catalytic residue, Glu86, is colored magenta. (c) SSV1 integrase (left, PDB ID 3VCF [36]) shares a conserved fold and catalytic residues with members belonging to the tyrosine recombinase family (highlighted in magenta), including the catalytic pentad (cluster of amino acids in the center of the protein) and the tyrosine nucleophile (distal to the active site). SIRV1 orf119 (right, PDB ID 2X3G [38]) has been identified as a novel type of Rep protein with the conserved fold of the superfamily II rep protein group. It shares conserved features with members of this group, including a conserved catalytic tyrosine (highlighted in magenta). Each subunit for the Rep protein dimer is colored differently.