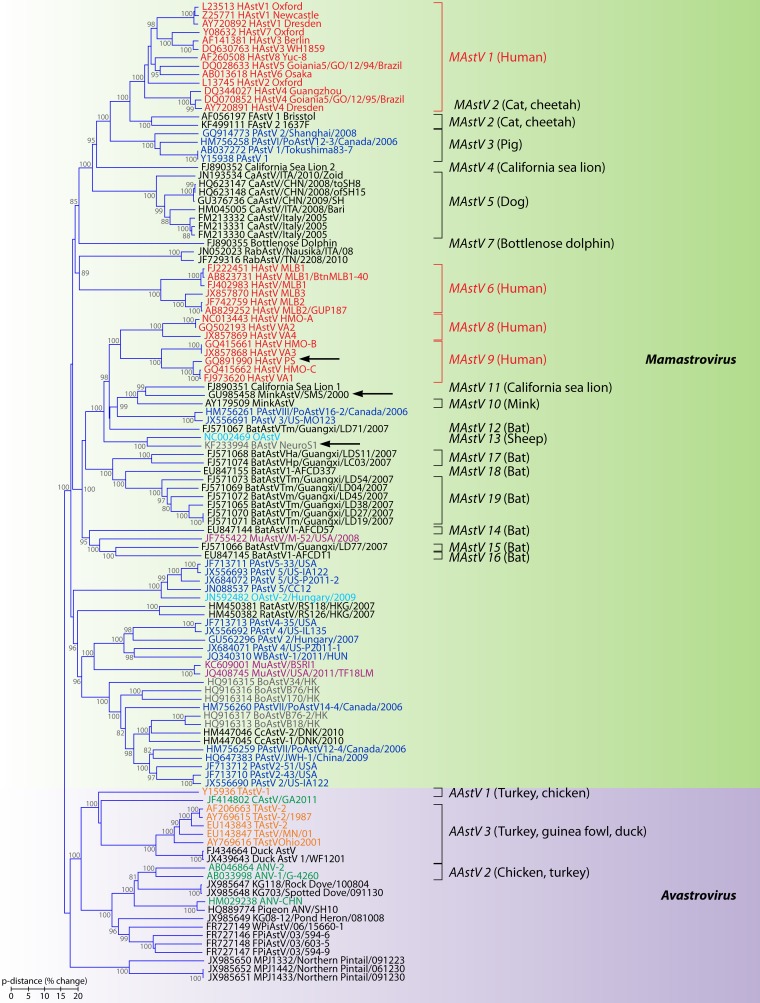
FIG 6.
Phylogenetic relationships within the family Astroviridae. The predicted amino acid sequences of the entire capsid polyprotein were aligned using Clustal Omega (309). The phylogenetic tree was generated using the neighbor-joining algorithm (310) implemented in the MEGA6 program (311). The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) which are higher than 70 are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances (p-dist) used to infer the phylogenetic tree. All positions containing alignment gaps and missing data were removed only in pairwise sequence comparisons (pairwise deletion option). Genotype species officially recognized by the ICTV are indicated, with hosts of origin shown in parentheses. Human viruses are colored in red, porcine viruses are colored in dark blue, mink viruses are colored in dark gray, bovine viruses are colored in brown, ovine viruses are colored in light blue, bat viruses are colored in black, murine viruses are colored in purple, turkey viruses are colored in orange, and chicken viruses are colored in green. Arrows indicate strains which have been isolated from neurologic tissues. HAstV, human astrovirus; FAstV, feline astrovirus; PAstV, porcine astrovirus; CaAstV, canine astrovirus; RabAstV, rabbit astrovirus; OAstV, ovine astrovirus; BoAstV, bovine astrovirus; MuAstV, murine astrovirus; WBAstV, wild boar astrovirus; CcAstV, deer astrovirus; TAstV, turkey astrovirus; CAstV, chicken astrovirus; ANV, avian nephritis virus; WPIAstV, wild pigeon astrovirus.