Abstract
Peptide-nucleotide antibiotic microcin C (McC) is produced by some Escherichia coli strains. Inside a sensitive cell, McC is processed, releasing a nonhydrolyzable analog of aspartyl-adenylate, which inhibits aspartyl-tRNA synthetase. The product of mccE, a gene from the plasmid-borne McC biosynthetic cluster, acetylates processed McC, converting it into a nontoxic compound. MccE is homologous to chromosomally encoded acetyltransferases RimI, RimJ, and RimL, which acetylate, correspondingly, the N termini of ribosomal proteins S18, S5, and L12. Here, we show that E. coli RimL, but not other Rim acetyltransferases, provides a basal level of resistance to McC and various toxic nonhydrolyzable aminoacyl adenylates. RimL acts by acetylating processed McC, which along with ribosomal protein L12 should be considered a natural RimL substrate. When overproduced, RimL also makes cells resistant to albomycin, an antibiotic that upon intracellular processing gives rise to a seryl-thioribosyl pyrimidine that targets seryl-tRNA synthetase. We further show that E. coli YhhY, a protein related to Rim acetyltransferases but without a known function, is also able to detoxify several nonhydrolyzable aminoacyl adenylates but not processed McC. We propose that RimL and YhhY protect bacteria from various toxic aminoacyl nucleotides, either exogenous or those generated inside the cell during normal metabolism.
INTRODUCTION
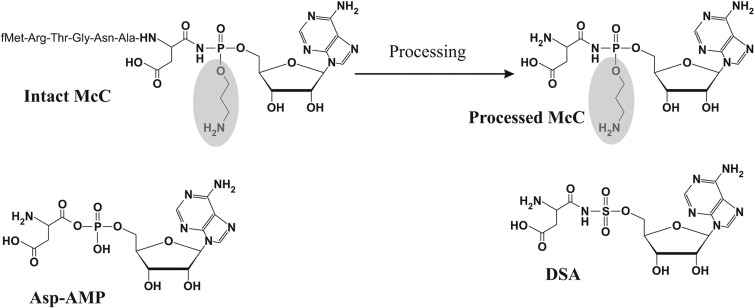
Peptide-nucleotide antibiotic microcin C (McC) consists of an MRTGNAD heptapeptide with a modified AMP attached to the α carboxyl group of the aspartate through an N-acyl phosphoramidate linkage (Fig. 1). The N-terminal methionine is formylated; the phosphate group is decorated with a propylamine group. The peptide moiety of McC is encoded by a 21-bp-long mccA gene (1, 2). The remaining genes of the mcc cluster are responsible for the synthesis of antibiotic (mccB, mccD, and mccE) and immunity of the producing cell to both endogenous and exogenous McC (mccC, mccE, and mccF).
FIG 1.
Microcin C and the mechanism of its action. The structures of intact McC, processed McC, aspartyl-adenylate, an intermediate of reaction catalyzed by AspRS, and aspartyl sulfamoyl adenylate DSA are shown. The propylamine group (shaded) is absent from McC(1120) and its processing product.
Escherichia coli cells actively import McC through the action of the YejABEF inner membrane transporter (3). Inside the cell, the N-terminal formyl group of McC is removed by peptide deformylase Pdf, and the peptide part is next degraded by one of the three cellular aminopeptidases—A, B, or N (4). As a result, processed McC is generated (Fig. 1). Processed McC is a nonhydrolyzable analog of aspartyl-adenylate (Fig. 1), an intermediate of a tRNAAsp aminoacylation reaction catalyzed by aspartyl-tRNA synthetase (AspRS) (5). Processed McC binds to AspRS and inhibits it, leading to the cessation of production of Asp-tRNAAsp, a stringent response, and the cessation of protein synthesis. Processed McC generated internally within the producing cell is detoxified by the acetyltransferase activity of the C-terminal domain of the MccE protein (MccECTD). MccECTD acetylates the primary amino group of the aminoacyl moiety of processed McC; acetylated processed McC no longer inhibits AspRS (6). MccECTD also acetylates (and, therefore, provides resistance to) a wide spectrum of nonhydrolyzable synthetic aminoacyl sulfamoyl adenylates (aaSAs, where “aa” denotes an amino acid in a single-letter code) such as DSA, shown in Fig. 1.
The MccE acetyltransferase is homologous to bacterial N-terminal acetyltransferases (NATs) of the Rim family. The E. coli genome encodes three Rim proteins, RimI, RimJ, and RimL, which acetylate ribosomal proteins S18, S5, and L12, respectively (7, 8). The physiological functions of these NATs and the significance of ribosomal protein acetylation for cell physiology are not entirely clear. Here, we investigated whether sequence similarity of MccECTD with Rim family NATs leads to functional similarity and allows Rim NATs to play a role in McC resistance. Our results indicate that RimL indeed contributes to the basal level of McC resistance through the same mechanism as the MccE acetyltransferase. In contrast, RimI and RimJ do not recognize processed McC or its analogs and play no role in McC resistance. We also demonstrate that an additional E. coli NAT, the product of the yhhY gene, acetylates some nonhydrolyzable aminoacyl adenylates (but not processed McC) and increases cellular resistance to these compounds.
MATERIALS AND METHODS
Bacterial strains and reagents.
E. coli NovaBlue Singles competent cells (Novagen) were used for cloning and plasmid propagation. E. coli BL21(DE3) isolates were used for recombinant protein overproduction. E. coli K-12 strain BW25113 [lacIp4000 (lacIq) rrnB3 ΔlacZ4787 hsdR541 Δ(araBAD)567 Δ(rhaBAD)568 rhp-1] (9) was used as a parental strain to generate rim knockouts. The YhhY open reading frame (ORF) clone from the ASKA library (10) was used for purification of the YhhY protein. All restriction enzymes, Vent DNA polymerase, Taq DNA polymerase, DNA, and protein molecular weight markers were purchased from New England BioLabs Inc. T4 DNA ligase was from Fermentas. All oligonucleotide primers were synthesized by Integrated DNA Technologies.
Microcin C and its maturation derivative McC(1120) were purified as described elsewhere (11). Albomycin was a gift of G. Katrukha (Gause Institute of Antibiotics, Moscow, Russia). Aminoacyl sulfamoyl adenylates and synthetic McC analogs were synthesized as described previously (12).
Cloning and protein expression.
The Nα-acetyltransferase genes—rimI, rimJ, and rimL—were amplified from E. coli BW25113 genomic DNA with appropriate primers containing engineered NdeI and EcoRI restriction sites for subsequent cloning. PCR products were blunt-end cloned into pT7Blue vector (Novagen), excised with NdeI and EcoRI enzymes, and recloned into pET28a expression vector (Novagen). All plasmids were confirmed by DNA sequencing.
E. coli BL21(DE3) cells harboring the pET28a-rimI (or -rimJ or -rimL) plasmids were grown in 1 liter of LB medium, supplemented with 25 μg/ml kanamycin, at 37°C until an optical density at 600 nm (OD600) of ∼1 was reached. Cultures were cooled down to 16°C, expression of plasmid-borne rim genes was induced by the addition of 0.2 mM isopropyl 1-thio-β-d-galactopyranoside, and growth at 16°C was continued for 20 to 24 h. Cells were harvested by centrifugation and stored at −80°C before use. An ASKA library clone overproducing YhhY was grown and induced according to a protocol published elsewhere (10).
In vivo sensitivity tests.
E. coli Bl21(DE3) cells carrying pET28a, pET28a-rimL, pET28a-rimI, pET28a-rimJ, or pET28a-yhhY plasmids were grown in 5 ml of LB medium supplemented with kanamycin (50 μg/ml) and isopropyl β-d-thiogalactopyranoside (0.1 mM) at 30°C overnight. One hundred microliters of culture was mixed with melted LB top (0.7 g/liter) agar and poured onto the surface of an LB agar plate supplemented with kanamycin (50 μg/ml). Drops (5 μl) of solutions of the antibiotics being tested were deposited on the solidified top agar layer. The concentration of McC, McC(1120), and albomycin was 100 μM, the concentration of synthetic McC analog solutions was 350 μM, and the concentration of aaSA solutions was 10 mM. Plates were incubated for several hours at 30°C. The sizes of growth inhibition zones around the drops of antibiotic solutions were monitored and recorded. Each experiment was repeated at least three times.
Tests with cells lacking rim genes were performed similarly, except that the LB medium contained no antibiotics and plates were incubated at 37°C.
Protein purification.
To purify RimI or RimL, the frozen cell mass was thawed and sonicated in a water-ice bath in buffer A containing 20 mM Tris-HCl (pH 8.0), 0.5 M NaCl, 2 mM 2-β-mercaptoethanol, and 0.1 mM phenylmethylsulfonyl fluoride (PMSF). The lysate was clarified by centrifugation for 30 min at 15,000 × g. The supernatant was supplemented with 5 mM imidazole and loaded onto a 5-ml Ni HiTrap column (GE Healthcare) equilibrated with buffer A containing 5 mM imidazole. Bound protein was step eluted with 20, 50, 100, and 200 mM imidazole in buffer A. Fractions containing pure Rim proteins (as revealed by Coomassie blue staining of SDS gels) were pooled and dialyzed against two exchanges of 500 volumes of 40 mM Tris-HCl (pH 8.0), 100 mM NaCl, 0.5 mM EDTA, 0.5 mM 2-β-mercaptoethanol, and 10% glycerol. Protein samples were supplemented with glycerol up to 50% stored at −20°C. To purify RimJ protein, a similar protocol was used but with pH 7.0 phosphate buffer instead of Tris buffer. The YhhY protein was purified on HIS-Select nickel affinity gel (Sigma) under denaturing conditions using 8 M urea for protein solubilization and chromatography according to the manufacturer's protocol. Fractions with pure protein were pooled and dialyzed against reconstitution buffer containing 40 mM Tris-HCl (pH 8.0), 200 mM NaCl, 0.5 mM EDTA, 0.5 mM 2-β-mercaptoethanol followed by dialysis against the storage buffer (reconstitution buffer containing 50% glycerol).
Preparation of E. coli S30 cell extracts.
E. coli cells were grown to an OD600 of ∼0.8 in LB medium. Cells were collected by centrifugation and washed with 40 mM Tris-HCl (pH 7.6), 10 mM Mg(OAc)2, 50 mM potassium acetate, 0.1 mM EDTA, and 1 mM dithiothreitol (DTT). The cell pellet was resuspended in an equal volume of the same buffer and disrupted using a French press (pressure, 108 Pa [1,000 bar]). The lysate was next centrifuged at 30,000 × g for 30 min. Then, supernatant was aliquoted and either directly used in tRNA aminoacylation reactions or stored at −80°C until further use.
tRNA aminoacylation reaction.
Aminoacylation reactions were carried out according to the method described in reference 5 with some modifications. To 1 μl of solution containing an inhibitor being tested, 3 μl of E. coli S30 extract was added. Next, 16 μl of aminoacylation mixture (30 mM Tris-HCl [pH 8.0], 1 mM DTT, 5 g/liter bulk E. coli tRNA [Sigma], 3 mM ATP, 30 mM KCl, 8 mM MgCl2, and 40 μM desired labeled amino acid) was added, and reaction mixtures were incubated at room temperature for 5 min. The reactions were terminated by the addition of cold 10% trichloroacetic acid (TCA), and precipitated reaction products were collected on Whatman 3MM paper filters. After thorough washing with cold 10% TCA, the filters were washed twice with acetone and dried on a heating plate. Following the addition of scintillation liquid, the amount of radioactivity was determined in a scintillation counter.
N-Acetyltransferase activity assay.
The N-acetyltransferase activity assay was performed in 20 μl of 100 mM Tris-HCl (pH 8.0), 0.1 mM EDTA, 1 mM acetyl coenzyme A (acetyl-CoA), and 0.5 to 1 mM substrate for 10 min at 25°C. Reactions were initiated by the addition of 0.07, 0.2, 1, and 2 μM purified recombinant YhhY, RimI, RimL, and RimJ, respectively, and were terminated by the addition of 500 μl of 0.05 mM 4,4′-dithiodipyridine (DTDP) (Aldrich) (13). Enzyme activity was determined by measuring the increase in absorbance at 324 nm due to the formation of 4-thiopyridone in the reaction between DTDP and the sulfhydryl group (SH) of the CoA-SH produced after the acetyl transfer reaction, and specific activity was calculated as the amount of substrate (μmol) converted into CoA-SH product by 1 mg of enzyme per minute of reaction.
To verify the acetyltransferase activity of recombinant Rim proteins, synthetic peptides (GenScript) matching the first 10 N-terminal residues of E. coli ribosomal proteins S18, S5, and L12 were used as the substrates (acetyl group acceptors) for RimI, RimJ (7), and RimL (8), respectively. Earlier, such an approach had been applied by the Blanchard group to analyze bisubstrate inhibition of RimI from Salmonella enterica serovar Typhimurium (14, 15) and during crystal structure determination of this enzyme (16). The recombinant enzyme preparations of RimI, RimJ, and RimL used in this work gave, respectively, the following specific activities toward chosen cognate synthetic substrates: 0.51, 0.021, and 0.052 μmol/min/mg. No activity to noncognate substrates was detected.
MALDI-TOF MS.
Samples for mass spectrometric (MS) analysis were dissolved in acetonitrile. Aliquots (0.5 μl) were mixed on a steel target with 1 μl of 2,5-dihydroxybenzoic acid (Aldrich) solution (20 mg/ml in 20% acetonitrile, 0.5% trifluoroacetic acid), and the droplet was left to dry at room temperature. The mass spectra were recorded on an ABI-MDS SCIEX 4800 matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometer. The [MH]+ molecular ions were measured in reflector and tandem (Lift) mode; the accuracy of mass peak measurement was 0.1 Da for parent ions and 0.5 Da for daughter ions.
RESULTS
Chromosomally encoded RimL contributes to the basal level of resistance to McC, its analogs, and nonhydrolyzable aminoacyl adenylates.
To determine the role of Rim NATs in McC resistance, E. coli K-12-based strains lacking individual rim genes were constructed. Three strains carrying double rim deletions (ΔrimI ΔrimJ, ΔrimI ΔrimL, and ΔrimJ ΔrimL) and a triple ΔrimI ΔrimJ ΔrimL mutant were also constructed. All strains were viable and grew equally well in rich medium at 37°C (data not shown). Susceptibilities of mutant strains to (i) McC, (ii) McC(1120), an 1,120-Da McC maturation intermediate lacking the propylamine modification (11), (iii) two aminoacyl sulfamoyl adenylates (aaSAs) active against E. coli (aspartyl sulfamoyl adenylate DSA and leucil sulfamoyl adenylate (LSA) (12), (iv) two synthetic McC(1120) analogs with an MRTGNA peptide (“X”) corresponding to the first six amino acids of McC heptapeptide coupled to DSA and LSA (12), and (v) albomycin were next determined. Drops of solutions containing various compounds were deposited on lawns of wild-type or mutant E. coli, and the sizes of growth inhibition zones were recorded after overnight incubation at 37°C. McC and its analogs were used at 100 and 350 μM concentrations. AaSAs, which lack the transport peptide, were tested at the much higher concentration of 10 mM in order to obtain observable growth inhibition zones. Albomycin was used at a 100 μM concentration. Results obtained with the triple mutant strain are shown in Fig. 2. As can be seen, the mutant cells were more sensitive to McC, its chemical analogs, or aaSAs than, but were as sensitive to albomycin as, the wild-type cells. The extent of protection afforded by chromosomal rim genes varied for different compounds. Protection was marginal for DSA but very obvious for McC and McC(1120).
FIG 2.
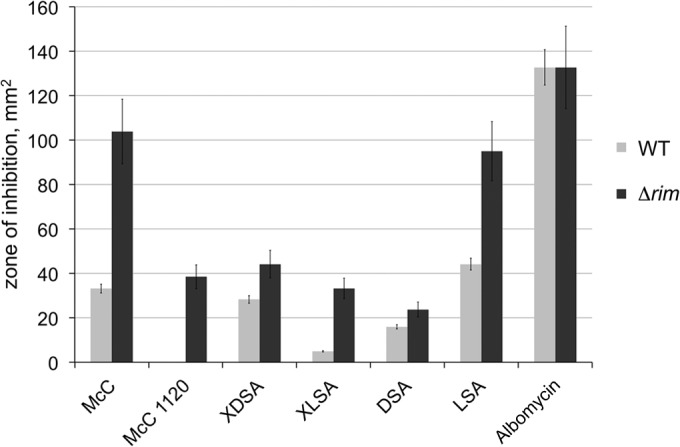
Cells lacking rim genes are hypersensitive to McC, its maturation intermediates, chemical analogs, and nonhydrolyzable aminoacyl adenylates but not to albomycin. Five-microliter drops of solutions containing the indicated compounds were deposited on lawns of wild-type or isogenic mutant ΔrimI ΔrimJ ΔrimL E. coli cells. After overnight growth at 37°C, the areas of growth inhibition zones were determined. The bars show mean values obtained in three independent experiments; the error bars show standard deviations. The compounds tested included the following: McC (a mature form with aminopropyl modification [Fig. 1]); McC(1120), an 1,120-Da McC maturation intermediate lacking the propylamine modification; XDSA and XLSA, which are chemically synthesized McC(1120) analogs containing an MRTGNA hexapeptide C-terminally fused to aspartyl or leucyl sulfamoyl adenylates, respectively; aminoacyl sulfamoyl adenylates DSA and LSA carrying aspartyl (Fig. 1) and leucyl aminoacyl moieties, respectively; and, finally, albomycin.
Deletions of rimI and/or rimJ had no effect on sensitivity to any of the compounds tested (data not shown). Further, the ΔrimI ΔrimL and ΔrimJ ΔrimL double mutants or the ΔrimI ΔrimJ ΔrimL triple mutant were no more sensitive than cells lacking rimL only (data not shown). The result thus suggests that RimL contributes to the basal level of E. coli resistance to McC, its analogues, and nonhydrolyzable aminoacyl-adenylates but not to albomycin. In contrast, RimL and RimI do not contribute to cellular resistance to any of the compounds tested.
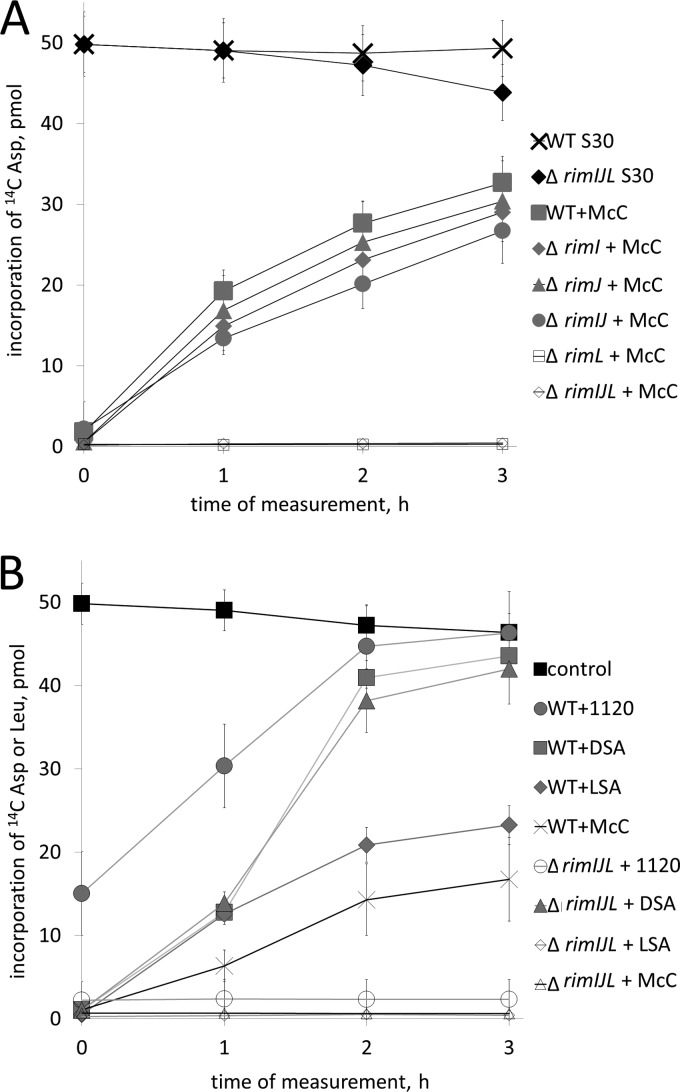
Cell extracts from wild-type and various rim mutant cells were prepared, supplemented with McC, and incubated for 15 min. Elsewhere, we show that a 15-min incubation is sufficient for complete processing of McC by cellular peptidases at the conditions of the assay (4). Indeed, the tRNAAsp aminoacylation reaction was strongly inhibited when measured after the processing was complete in all extracts (Fig. 3A). However, the tRNAAsp aminoacylation activity was partially recovered in extracts of wild-type cells that were incubated with McC for longer periods of time (Fig. 3A). The recovery of tRNAAsp aminoacylation activity in extracts prepared from ΔrimI, ΔrimJ, or ΔrimI ΔrimJ cells closely followed that observed in the wild-type cell extract (Fig. 3A). In contrast, no such recovery was observed in extracts prepared from ΔrimL, ΔrimI ΔrimL or ΔrimJ ΔrimL double mutants or from ΔrimI ΔrimJ ΔrimL triple mutant cells: the tRNAAsp aminoacylation activity in these extracts appeared to be inhibited irreversibly (Fig. 3A). The results thus indicate that cellular RimL contributes to the basal level of E. coli resistance to McC by inactivating processed McC.
FIG 3.
RimL detoxifies processed McC and LSA in vitro. (A) AspRS-catalyzed aminoacylation of tRNAAsp in S30 extracts prepared from wild-type E. coli and the indicated rim mutants. Extracts were supplied with intact McC and incubated for 15 min to allow processing. Aliquots of extracts were removed, and tRNAAsp aminoacylation reactions were carried out. The first aliquots were removed right after the initial 15-min incubation needed for McC processing. This time point is labeled “0.” The amounts of aminoacylated tRNAAsp (measured as incorporation of [C14] Asp in TCA-precipitable material) are shown. Control shows the time course of aminoacylation levels in wild-type S30 cell extracts without any additions. Data from three independently performed experiments (mean values and standard deviations) are shown. (B) E. coli S30 extracts prepared from wild-type or a triple rim deletion E. coli strain were combined with intact McC, McC(1120), DSA, or LSA. Extracts containing McC or McC(1120) were incubated for 15 min to allow processing before initiating tests for tRNAAsp aminoacylation (times are indicated as indicated for panel A); in extracts containing DSA or LSA, measurements of tRNAAsp or tRNALeu aminoacylation, respectively, were initiated immediately after the addition of inhibitors. Data from three independently performed experiments (mean values and standard deviations) are shown.
The addition of McC(1120) to wild-type cell extract led to partial inhibition of tRNAAsp aminoacylation at the time of the first measurement after a 15-min preincubation to allow processing (Fig. 3B). Upon further incubation, complete recovery from inhibition by McC(1120) was observed. In extracts of triple mutant cells, McC(1120) fully inhibited tRNAAsp aminoacylation without subsequent recovery of AspRS activity (Fig. 3B). We conclude that RimL inactivates processed McC(1120) more efficiently than processed McC. The incomplete initial inhibition by McC(1120) in wild-type cell extract is therefore likely due to a partial detoxification of processed McC(1120) that must be occurring simultaneously with the processing of intact McC(1120) during the initial 15-min incubation.
The effect of the DSA addition on aminoacylation of tRNAAsp in extracts from wild-type or triple mutant cells was also tested. Since DSA does not require processing, the first measurement of AspRS activity was performed immediately after the addition of DSA. As can be seen (Fig. 3B), the AspRS activity in both extracts was initially inhibited by the addition of DSA but eventually completely recovered from the inhibition. The absence of requirement for Rim proteins for recovery from DSA inhibition agrees with in vivo data showing that the contribution of intracellular Rim proteins to DSA (or XDSA) resistance is only slight (Fig. 2). LSA, a sulfamoyl adenosine inhibitor of LeuRS, irreversibly inhibited aminoacylation of tRNALeu in triple mutant cell extracts, but partial recovery from inhibition was observed in wild-type cell extracts (Fig. 3B), again in agreement with in vivo data, which showed that cells lacking rimL were significantly more sensitive to LSA than wild-type cells (Fig. 2).
Effects of Rim proteins on resistance to McC and related compounds.
Our results suggest either that RimI and RimJ are not necessary for detoxification of McC and toxic aaSAs or that intracellular amounts of these proteins are too low to affect McC and aaSA resistance. PET-based expression plasmids overproducing E. coli RimI, RimJ, or RimL were constructed. SDS-PAGE analysis indicated that all three proteins were overproduced at high (and similar) levels (data not shown). Sensitivity of E. coli BL21(DE3) cells harboring Rim-overproducing plasmids to McC, a collection of bioactive aaSAs (DSA, LSA, KSA, ASA, GSA, FSA, and PSA), and albomycin was tested. We used all bioactive aaSAs available to us since we considered that RimI and/or RimJ may be specific for certain aminoacyl adenylates. Previously, we showed that cells overproducing MccECTD are resistant to McC, DSA, LSA, KSA, ASA, GSA, FSA, and albomycin but are sensitive to PSA (6). This result was reproduced (Table 1). Cells overproducing RimL were indistinguishable from MccECTD-overproducing cells (Table 1). In contrast, cells overproducing RimI or RimJ were as sensitive as cells harboring the PET vector plasmid to every compound tested (Table 1).
TABLE 1.
Sensitivity of cells overproducing Rim proteins and YhhY to McC, albomycin, and aaSAsa
| Protein | McC | Albomycin | DSA | LSA | KSA | GSA | ASA | FSA | PSA |
|---|---|---|---|---|---|---|---|---|---|
| RimL | R | R | R | R | R | R | R | R | S |
| RimI | S | S | S | S | S | S | S | S | S |
| RimJ | S | S | S | S | S | S | S | S | S |
| YhhY | S | S | S | R | S | S | PR | PR | S |
R, absence of visible growth inhibition zones at the concentration of inhibitor used; S, clear growth inhibition zones of the same size as observed on control cell lawn formed by cells carrying a pET28a plasmid vector; PR (partial resistance), presence of turbid growth inhibition zones whose diameter was at least twice less than that on control lawn.
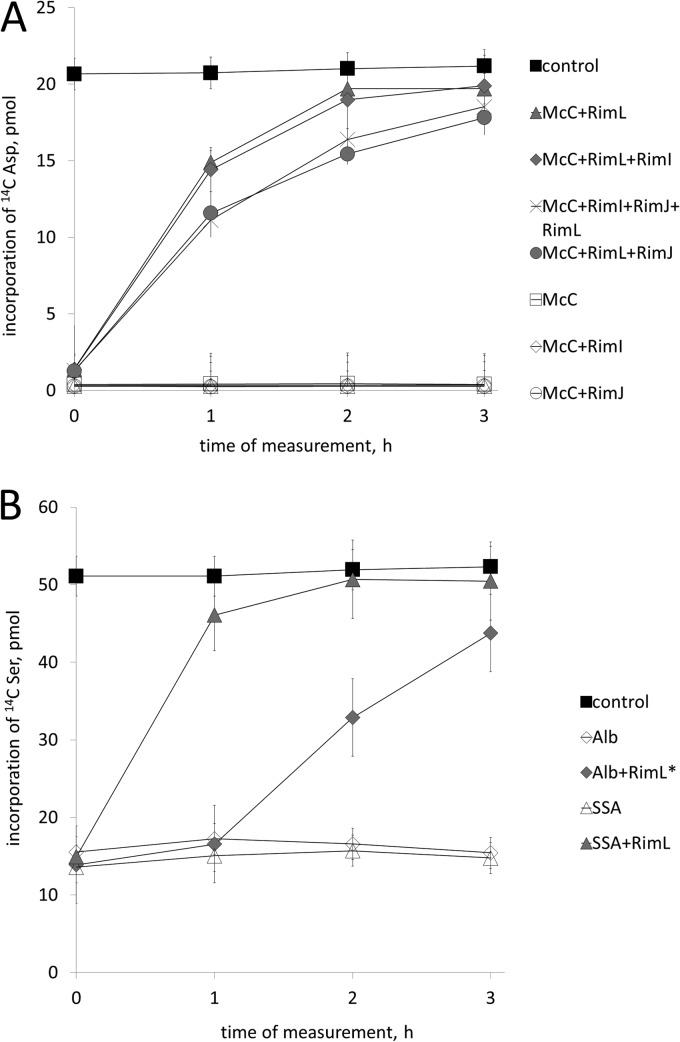
The inability of overproduced RimI or RimJ to provide resistance to McC could have been due to poor solubility and/or folding of recombinant proteins. To rule out this possibility, recombinant Rim proteins were purified from cytoplasmic extracts of appropriate overproducing cells to apparent homogeneity. Control experiments established that each protein was acetylating its cognate substrates and was therefore properly folded and active (see Materials and Methods). The addition of recombinant MccECTD or RimL—but not RimI or RimJ—to extracts of ΔrimI ΔrimJ ΔrimL cells inhibited with McC led to almost complete recovery of tRNAAsp aminoacylation after a 3-hour incubation (Fig. 4A). Simultaneous addition of RimI and RimJ had no effect, and neither RimI nor RimJ stimulated RimL-mediated recovery of tRNAAsp aminoacylation (Fig. 4A).
FIG 4.
RimL protein is sufficient for McC, McC(1120), DSA, XDSA, LSA, and albomycin resistance in vitro. Wild-type E. coli S30 extracts were supplemented with McC (A) or albomycin (B) and incubated to allow processing. Next, the indicated purified Rim proteins were added to reaction mixtures (time “0”), and incubation was continued. At various time points after addition of Rim proteins, reaction aliquots were withdrawn and tRNAAsp (A) or tRNASer (B) aminoacylation reactions were carried out. In panel B, a 20-fold-higher concentration of RimL (labeled with an asterisk) was used in reaction mixtures containing albomycin than in those containing SSA, which was used as a control. Data from three independently performed experiments with each inhibitor are shown (with means and standard deviations).
The addition of RimL to albomycin-inhibited extracts led to recovery of tRNASer aminoacylation (Fig. 4B). No such effect was observed with RimI or RimJ (data not shown). Since ∼20-fold-larger amounts of RimL were required to recover tRNASer aminoacylation in albomycin-inhibited extracts than to achieve full recovery of tRNAAsp aminoacylation in McC-inhibited extracts, processed albomycin is a much poorer RimL substrate than processed McC, in agreement with in vivo data (Fig. 2) and earlier findings with MccECTD (6).
Purified Rim proteins were combined with acetyl-CoA, a donor of acetyl groups, and the rate of production of free CoA generated during the acetylation reaction was determined in the presence of processed McC, DSA, ESA, and LSA as described in Materials and Methods. The results, presented in Table 2, demonstrate that production of CoA from acetyl-CoA was observed with every aaSA tested in the presence of RimL. A similar result was obtained in the presence of MccECTD, as expected (6). In contrast, no CoA production was detected in reaction mixtures that contained RimI or RimJ.
TABLE 2.
In vitro substrate specificity of E. coli Rim and YhhY acetyltransferases
| Substrate | Sp acta (μmol/mg/min) |
|||
|---|---|---|---|---|
| RimI | RimJ | RimL | YhhY | |
| DSA | 0 | 0 | 2.39 | 0 |
| ESA | 0 | 0 | 1.54 | 0 |
| LSA | 0 | 0 | 2.11 | 28 |
| ISA | ND | ND | 2.5 | 17 |
| Albomycin | 0 | 0 | 0 | ND |
Amount of product in the presence of 1 mM acetyl-CoA and 0.5 to 10 mM substrates (see Materials and Methods); ND, not determined.
RimL-catalyzed acetylation of aaSAs was confirmed by MALDI-TOF MS analysis of reaction products (Fig. 5 and data not shown). In all cases, a mass peak corresponding to the reaction substrate disappeared upon incubation with pure RimL and acetyl-CoA. Instead, a new peak whose m/z value was increased by 42 atomic units, corresponding to a transfer of an acetyl group, was observed. Thus, just like MccECTD, RimL can utilize aaSAs as the substrates for acetylation with little specificity with respect to the amino acid attached to the nucleotide moiety. RimI and RimJ are inactive in this reaction.
FIG 5.
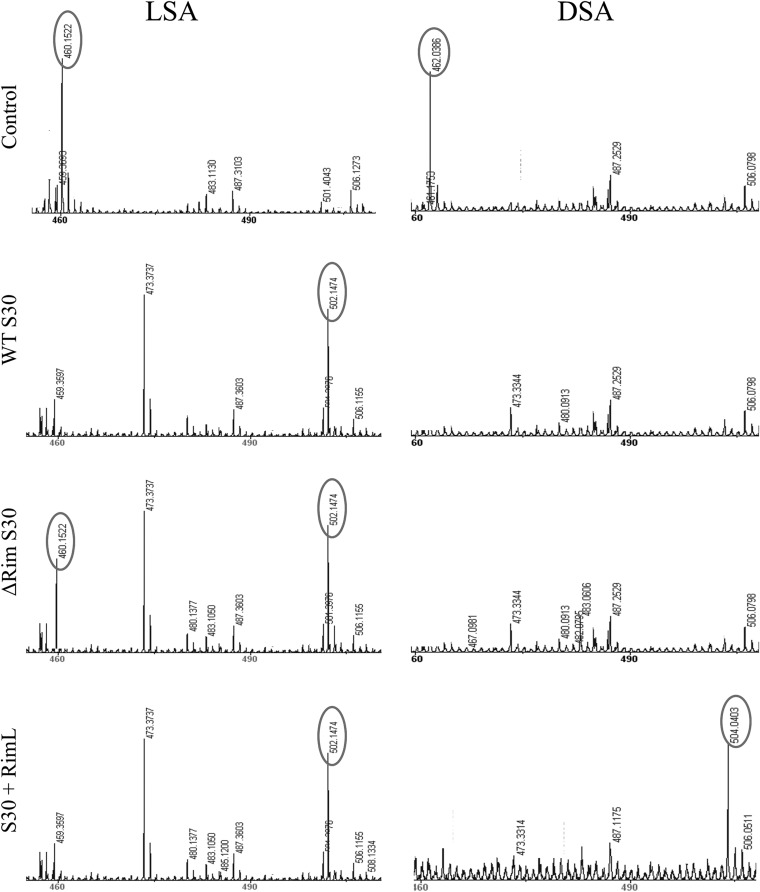
Mass spectrometric analysis of LSA and DSA incubated in various cell extracts. LSA and DSA were incubated with a buffer (control) or with S30 extracts prepared from wild-type, Δrim, or RimL-overproducing cells, and reactions were subjected to MALDI-TOF MS analysis. Only relevant portions of mass spectra are shown. The peaks corresponding to mass ions described in the text are in ovals.
Identification of an additional E. coli NAT capable of acetylating aminoacyl adenylates.
MALDI-TOF MS analysis of wild-type cells extract that recovered from LSA inhibition revealed that a mass peak corresponding to LSA (m/z = 460) disappeared. Instead, a peak corresponding to acetylated LSA (m/z = 502) was observed (Fig. 5). Interestingly, incubation in triple rim mutant cell extracts also led to the accumulation of detectable amounts of acetylated form of LSA, though the peak corresponding to unmodified LSA was also present (Fig. 5). Incubation of wild-type or rim mutant cell extracts with DSA led to the disappearance of the mass peak with an m/z of 462, corresponding to DSA (Fig. 5). No peak corresponding to acetylated or any other modified form of DSA was observed. The lack of accumulation of acetylated DSA was not caused by the inability of RimL to acetylate DSA, since acetylation products were readily detected upon incubation of either DSA or LSA with extract prepared from RimL-overproducing cells (Fig. 5). The results thus suggest that in wild-type cells DSA is efficiently degraded in a RimL-independent way, in agreement with results of in vivo sensitivity tests (Fig. 2) and in vitro results (Fig. 3B).
Accumulation of a +42 peak in LSA-inhibited extracts lacking Rim proteins suggested that there exists yet another, non-Rim, NAT that can acetylate LSA. Bioinformatics analysis of the E. coli genome reveals several genes coding for putative or experimentally proven acetyltransferases different from the Rim proteins. The products of two such genes, yhhY and speG, are most closely related to Rim proteins (Fig. 6A) and were investigated further. A yhhY deletion strain, a quadruple mutant strain lacking the three rim genes and the yhhY gene, and a yhhY expression plasmid were generated. The corresponding speG deletion strains and an expression plasmid were also made. The sensitivity of yhhY or speG deletion strains to various inhibitors used in the experiments described above was not different from those of control (wild-type or triple rim mutant) strains (data not shown). SpeG overproduction had no effect on the sensitivity of cells to McC and aaSAs (data not shown). In contrast, YhhY-overproducing cells became completely resistant to LSA and were partially resistant to ASA and FSA but remained as sensitive to McC, DSA, KSA, GSA, and PSA as control cells harboring an empty vector plasmid (Table 1). The results thus indicate that YhhY, when overproduced, can indeed provide resistance to some nonhydrolyzable aminoacyl adenylates.
FIG 6.
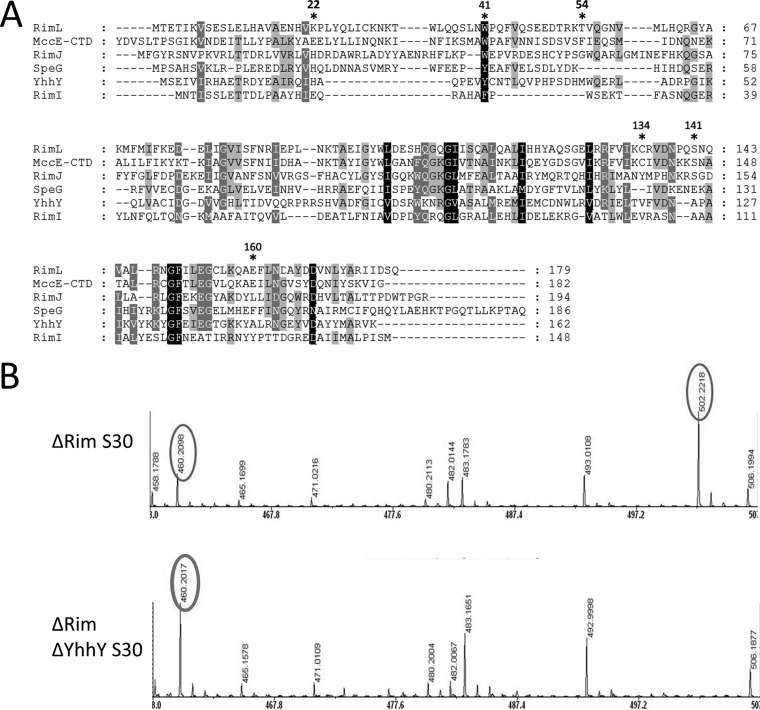
YhhY is an acetyltransferase that detoxifies nonhydrolyzable aminoacyl adenylates in vivo and in vitro. (A) A multiple sequence alignment of E. coli Rim proteins, MccECTD, SpeG, and the YhhY protein. The extent of sequence conservation is indicated by background shading. Catalytic and substrate binding residues are marked with asterisks. Residues 22, 41, and 54 of RimL correspond to known adenine-binding residues of MccECTD and likely adenine-binding residues of YhhY: RimL Lys22 corresponds to His21 of YhhY; Trp41 corresponds to Trp453 of MccE (Trp46 of MccECTD) and Trp41 of YhhY, and Thr54 corresponds to Phe466 of MccE (Phe59 of MccECTD) (17). Residue Cys134 of RimL forms a disulfide bond with acetyl-CoA; Ser141 and Glu160 act as general acid/base catalysts of the acetyltransferase reaction. Alignment was built using MUSCLE software, and the figure was prepared using GENEDOC. (B) LSA was incubated with extracts prepared from indicated cells and subjected to MALDI-MS analysis. Only relevant portions of mass spectra are shown.
S30 extracts from triple rim and quadruple deletion (triple rim and yhhY) were incubated with LSA, followed by MALDI-TOF MS analysis. While partial conversion of LSA to a +42 mass ion with an m/z of 502 was detected in triple mutant cells, the LSA peak remained intact in the quadruple mutant cell extract and no peak with m/z of 502 was detected (Fig. 6B). We therefore conclude that endogenous YhhY is responsible for acetylation of LSA in the absence of RimL.
The YhhY protein was purified, and its ability to acetylate several substrates was determined. The results, presented in Table 2, indicate that YhhY was approximately 10 times more active than RimL in acetylation of LSA and ISA but was unable to utilize DSA and ESA as acetylation substrates, in agreement with in vivo results.
DISCUSSION
The principal result of this work is the demonstration that the Escherichia coli RimL protein contributes to microcin C resistance by acetylating the amino group of the processed McC aspartate moiety. In this respect, RimL appears to be very similar to the C-terminal domain of MccE acetyltransferase encoded by the plasmid-borne McC biosynthesis/immunity gene cluster mcc (6). As is the case with MccECTD, the nature of the amino acid appears to be unimportant for RimL activity, since all aminoacyl sulfamoyl adenylates containing the primary amine group tested in this work (i.e., Ala, Asp, Gly, Phe, Lys, Glu, Leu, and Ile sulfamoyl adenylates) are efficiently acetylated. The nature of the bond between the aminoacyl and nucleotide moiety is also unimportant, since compounds containing either phosphoamide or sulfamoyl are recognized well. Processed albomycin, which contains a pyrimidine nucleotide instead of adenosine, is also acetylated by RimL, albeit much less efficiently. Like the MccE acetyltransferase, RimL does not use free amino acids or intact McC as the substrates.
Structural analysis indicates that the promiscuity of MccECTD with respect to aminoacyl moieties of its substrates is due to tight binding to substrate nucleotidyl moiety through stacking interactions with two aromatic amino acids of the protein, Trp453 and Phe466, and the absence of contacts with the aminoacyl moiety of the substrate (17). The available crystal structure of RimL from Salmonella (16) reveals a conservation of only one of the π-stacking residues, Trp41, which corresponds to MccE Trp453 (MccECTD Trp46) (Fig. 6A). Though a second aromatic residue corresponding to MccE Phe466 (MccECTD Phe59) is missing, the other face of the active site of RimL is lined with hydrophobic amino acids such as Leu24, Met69, and Val81; a histidine side chain (His54) is also placed in close proximity. In the Salmonella RimI crystal structure (18), a duet of similar stacking residues—Trp27 and His32—can be identified. However, the adenine-binding site in RimI is occluded by the carboxylate of Glu19 and the amide side chain of Asn68. Steric occlusion by these residues could contribute to the observed inability of RimI to acetylate aminoacyl adenylates by preventing their binding.
In the active site of YhhY predicted by alignment with the MccECTD structure, His21 and the indole ring of Trp41 are predicted to stack on either side of the substrate adenine ring analogous to Trp453 and Phe466 of MccECTD. Thus, a purine nucleobase binding motif created by two parallel planar aromatic amino acid side chains appears to be a common mechanism for engagement of aminoacyl adenylates by NATs. The narrower specificity of YhhY toward its substrates (the obvious preference for compounds with amino acids containing nonpolar side chains) suggests that this protein contains an additional hydrophobic pocket that accepts this part of the substrate.
Interestingly, our data reveal that though purified RimL can effectively acetylate and therefore detoxify DSA, another system efficiently removes DSA from E. coli cell extracts whether RimL is present or not. This yet-to-be identified system appears to degrade DSA, which completely disappears from the extracts, and mass spectrometric analysis fails to reveal peaks that could correspond to modified DSA. This additional system is not functional against McC(1120), whose processing product is identical for DSA except for a phosphoamide bond instead of sulfamoyl.
There are three Rim proteins encoded by the E. coli genome. RimI was discovered as an enzyme responsible for acetylation of ribosomal protein S18 (19), and RimJ acetylates ribosomal protein S5 (20), while RimL acetylates the L12 protein, converting it to L7 (21). The importance of the Rim proteins and the acetyl transfer reactions that they catalyze for cell physiology is not clear at present. In vitro, L7 forms a more stable complex with the L10 protein than does L12 (22). The L7/L12 ratio is higher in cells grown in poor medium (23), suggesting that RimL activity is activated by nutrient deprivation. These conditions also stimulate McC production by cells harboring the mcc operon (24). Be that as it may, none of the Rim proteins or combinations thereof is essential for viability at 37°C (though temperature sensitivity was noted for a rimL rimJ deletion mutant [18–20]). In fact, Rim proteins may have functions that are unrelated to N-acetylase activity. For example, RimJ plays an important role in the process of assembly of the 30S ribosomal subunits (25), and this involvement does not depend on the function of acetyltransferase. The data presented here indicate that some Rim proteins and other E. coli NATs of unassigned functions may acetylate aminoacyl nucleotides that accumulate either during cell metabolism or as a result of antibiotic treatment and thus contribute to bacterial cell fitness. Moreover, these proteins can also serve as an important source of antibiotic resistance.
ACKNOWLEDGMENTS
This work was supported by the U54 AI057158 NIH/NIAID North Eastern Biodefense Center grant, Russian Academy of Sciences Presidium programs in Molecular and Cellular Biology and in Nanotechnology, the Ministry of Education and Science of the Russian Federation project 14.B25.31.0004 grant, a Rutgers University Technology Commercialization Fund grant (to K.S.), and a Russian Foundation for Basic Research grant (06-04-48865) (to A.M.). G.H.V. is a recipient of a Belgian IWT fellowship.
Footnotes
Published ahead of print 7 July 2014
REFERENCES
- 1.Gonzalez-Pastor JE, San Millan JL, Moreno F. 1994. The smallest known gene. Nature 369:281–281. 10.1038/369281a0 [DOI] [PubMed] [Google Scholar]
- 2.Fomenko DE, Metlitskaya AZ, Peduzzi J, Goulard C, Katrukha GS, Gening LV, Rebuffat S, Khmel IA. 2003. Microcin C51 plasmid genes: possible source of horizontal gene transfer. Antimicrob. Agents Chemother. 47:2868–2874. 10.1128/AAC.47.9.2868-2874.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Novikova M, Metlitskaya A, Datsenko K, Kazakov T, Kazakov A, Wanner B, Severinov K. 2007. The E. coli Yej ABC transporter is required for the uptake of translation inhibitor microcin C. J. Bacteriol. 189:8361–8365. 10.1128/JB.01028-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kazakov T, Vondenhoff GH, Datsenko KA, Novikova M, Metlitskaya A, Wanner BL, Severinov K. 2008. E. coli peptidases A, B, or N can process translation inhibitor microcin C. J. Bacteriol. 190:2607–2610. 10.1128/JB.01956-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Metlitskaya A, Kazakov T, Kommer A, Pavlova O, Praetorius-Ibba M, Ibba M, Krasheninnikov I, Kolb V, Khmel I, Severinov K. 2006. Aspartyl-tRNA synthetase is the target of peptidenucleotide antibiotic Microcin C. J. Biol. Chem. 281:18033–18042. 10.1074/jbc.M513174200 [DOI] [PubMed] [Google Scholar]
- 6.Novikova M, Kazakov T, Vondenhoff GH, Semenova E, Rozenski J, Metlytskaya A, Zukher I, Tikhonov A, Van Aerschot A, Severinov K. 2010. MccE provides resistance to protein synthesis inhibitor microcin C by acetylating the processed form of the antibiotic. J. Biol. Chem. 285:12662–12669. 10.1074/jbc.M109.080192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshikawa A, Isono S, Sheback A, Isono K. 1987. Cloning and nucleotide sequencing of the genes rimI and rimJ which encode enzymes acetylating ribosomal proteins S18 and S5 of Escherichia coli K12. Mol. Gen. Genet. 209:481–488. 10.1007/BF00331153 [DOI] [PubMed] [Google Scholar]
- 8.Tanaka S, Matsushita Y, Yoshikawa A, Isono K. 1989. Cloning and molecular characterization of the gene rimL which encodes an enzyme acetylating ribosomal protein L12 of Escherichia coli K12. Mol. Gen. Genet. 217:289–293. 10.1007/BF02464895 [DOI] [PubMed] [Google Scholar]
- 9.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. 2005. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 12:291–299 [DOI] [PubMed] [Google Scholar]
- 11.Metlitskaya A, Kazakov T, Vondenhoff GH, Novikova M, Shashkov A, Zatsepin T, Semenova E, Zaitseva N, Ramensky V, Van Aerschot A, Severinov K. 2009. Maturation of the translation inhibitor microcin C. J. Bacteriol. 191:2380–2387. 10.1128/JB.00999-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van de Vijver P, Vondenhoff GH, Kazakov TS, Semenova E, Kuznedelov K, Metlitskaya A, Van Aerschot A, Severinov K. 2009. Synthetic microcin C analogs targeting different aminoacyl-tRNA synthetases. J. Bacteriol. 191:6273–6280. 10.1128/JB.00829-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abo-Dalo B, Ndjonka D, Pinnen F, Liebau E, Luersen K. 2004. A novel member of the GCN5-related N-acetyltransferase superfamily from Caenorhabditis elegans preferentially catalyses the N-acetylation of thialysine [S-(2-aminoethyl)-L-cysteine]. Biochem. J. 384:129–137. 10.1042/BJ20040789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu M, Magalhães ML, Cook PF, Blanchard JS. 2006. Bisubstrate inhibition: theory and application to N-acetyltransferases. Biochemistry 45:14788–14794. 10.1021/bi061621t [DOI] [PubMed] [Google Scholar]
- 15.Magalhães ML, Vetting MW, Gao F, Freiburger L, Auclair K, Blanchard JS. 2008. Kinetic and structural analysis of bisubstrate inhibition of the Salmonella enterica aminoglycoside 6′-N-acetyltransferase. Biochemistry 47:579–584. 10.1021/bi701957c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vetting MW, de Carvalho LPS, Roderick SL, Blanchard JS. 2005. A novel dimeric structure of the RimL Nalpha-acetyltransferase from Salmonella typhimurium. J. Biol. Chem. 280:22108–22114. 10.1074/jbc.M502401200 [DOI] [PubMed] [Google Scholar]
- 17.Agarwal V, Metlitskaya A, Severinov K, Nair SK. 2011. Structural basis for microcin C7 inactivation by the MccE acetyltransferase. J. Biol. Chem. 286:21295–21303. 10.1074/jbc.M111.226282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vetting MW, Bareich DC, Yu M, Blanchard JS. 2008. Crystal structure of RimI from Salmonella typhimurium LT2, the GNAT responsible for N(alpha)-acetylation of ribosomal protein S18. Protein Sci. 17:1781–1790. 10.1110/ps.035899.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isono K, Isono S. 1980. Ribosomal protein modification in Escherichia coli. II. Studies of a mutant lacking the N-terminal acetylation of protein S18. Mol. Gen. Genet. 177:645–651 [DOI] [PubMed] [Google Scholar]
- 20.Cumberlidge AG, Isono K. 1979. Ribosomal protein modification in Escherichia coli. I. A mutant lacking the N-terminal acetylation of protein S5 exhibits thermosensitivity. J. Mol. Biol. 131:169–189 [DOI] [PubMed] [Google Scholar]
- 21.Isono S, Isono K. 1981. Ribosomal protein modification in Escherichia coli. III. Studies of mutants lacking an acetylase activity specific for protein L12. Mol. Gen. Genet. 183:473–477 [DOI] [PubMed] [Google Scholar]
- 22.Gordiyenko Y, Deroo S, Zhou M, Videler H, Robinson CV. 2008. Acetylation of L12 increases interactions in the Escherichia coli ribosomal stalk complex. J. Mol. Biol. 380:404–414. 10.1016/j.jmb.2008.04.067 [DOI] [PubMed] [Google Scholar]
- 23.Deusser E, Wittmann HG. 1972. Ribosomal proteins: variation of the protein composition in Escherichia coli ribosomes as function of growth rate. Nature 238:269–270. 10.1038/238269a0 [DOI] [PubMed] [Google Scholar]
- 24.Diaz-Guerra L, Moreno F, San Millan JL. 1989. appR gene product activates transcription of microcin C7 plasmid genes. J. Bacteriol. 171:2906–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roy-Chaudhuri B, Kirthi N, Kelley T, Culver GM. 2008. Suppression of a cold-sensitive mutation in ribosomal protein S5 reveals a role for RimJ in ribosome biogenesis. Mol. Microbiol. 68:1547–1559. 10.1111/j.1365-2958.2008.06252.x [DOI] [PMC free article] [PubMed] [Google Scholar]