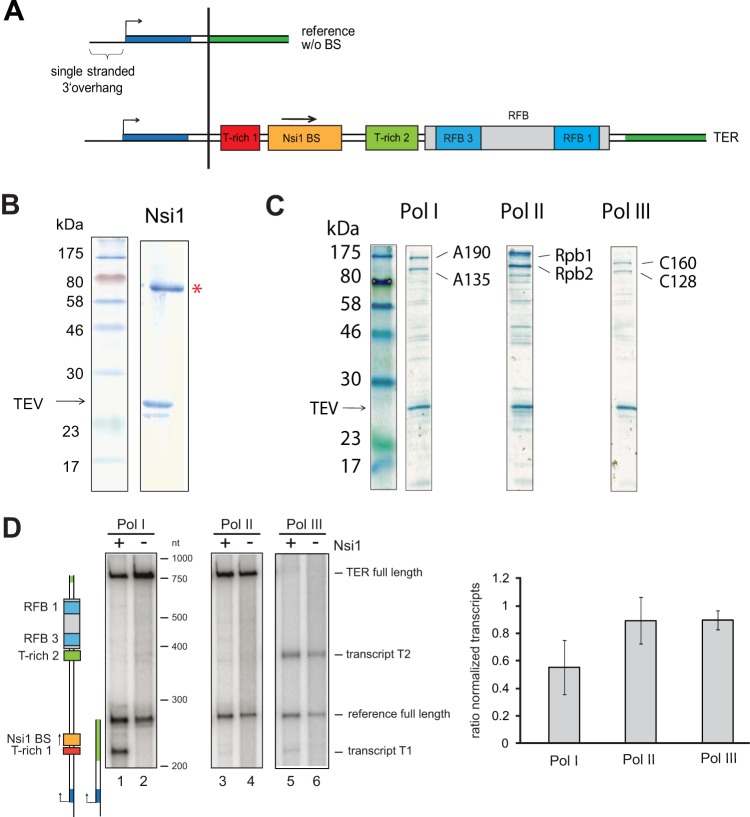
FIG 1.
Nsi1 pauses efficiently Pol I transcription at the Nsi1 binding site in tailed-template transcription assays. (A) Schematic representation of the DNA matrices for tailed-template assays. Transcription is initiated on a 24-nt single-stranded 3′ extension of the template strain. The transcription start site is indicated with a black arrow. Yeast 35S rDNA terminator region cis elements are depicted with red (T-rich 1), orange (Nsi1 binding site), and light green (T-rich 2) boxes. Two cyan boxes embedded in a gray rectangle represent the RFB 1 and RFB 3 regions within the replication fork barrier. The DNA stretches shown in dark blue and dark green of the reference template and of template TER denote sequence identity of the respective template sections. TER refers to the region from bp +70 to bp +414 with respect to the 3′ end of the 25S rDNA and with the Nsi1 binding site stretching from +109 to +119. Note that the DNA sequence coding for the Rnt1 cleavage site is not included. The reference template (254 nt) contains the same 3′ extension to start transcription but no DNA sequence from the terminator region. (B) Coomassie-stained gel of recombinant Nsi1 purified from Sf21 cells via FLAG immunoprecipitation (see Materials and Methods). Ten percent of the eluate derived from lysates of 50 × 106 infected cells was separated on an SDS gel. A red asterisk marks the band representing Nsi1, and an arrow indicates TEV protease. (C) Coomassie-stained gel of affinity-purified yeast RNA Pol I, II, and III. Pol I, II, and III were purified from yeast strain y2423, y2424, and y2425 in which subunits A135, Rpb2, and C128 are expressed with C-terminal protein A tags, respectively (see Materials and Methods). Ten percent of affinity-purified Pol I/II/III from a cell lysate representing an 800-ml culture volume at an OD600 of 1.5 was loaded on an SDS gel. Bands representing the two large subunits of each polymerase and TEV protease are indicated (see also Fig. S2 in the supplemental material for comparative mass spectrometric analysis). (D) In vitro transcription reactions were carried out in the presence of a template carrying the complete 35S rDNA terminator region (TER) and the reference template which did not contain putative termination elements. Experiments were conducted with one-step purified Pol I, II, and III. Final template concentration was 10 nM each. Nsi1 was preincubated with the template (+) and the nucleoside triphosphates for 15 min at room temperature before the reaction was started by addition of 7.5 nM RNA Pol I. Nsi1 concentration was 88 nM. Control reactions (−) were supplemented with Nsi1 storage buffer and processed equally. Transcription was stopped after 30 min by addition of proteinase K. The RNAs were extracted and separated on a denaturing polyacrylamide urea gel, and the radiolabeled transcripts were visualized side-by-side with an RNA marker which was in vitro transcribed with T7 RNA polymerase. Cartoons visualize the length and identity of the in vitro-transcribed RNAs. For quantification, signal intensities of TER full-length transcripts (757 nt) were normalized to reference full-length transcript (254 nt) signal intensities in the respective lanes. Then, the ratio of the normalized TER full-length transcript signal intensities in presence (+) versus absence (−) of Nsi1 was calculated. Experiments were conducted at least in duplicate, and mean values were plotted. Error bars represent ±1 standard deviation of the sample. (see Fig. S1 in the supplemental material for template specific transcription reactions).