FIG 5.
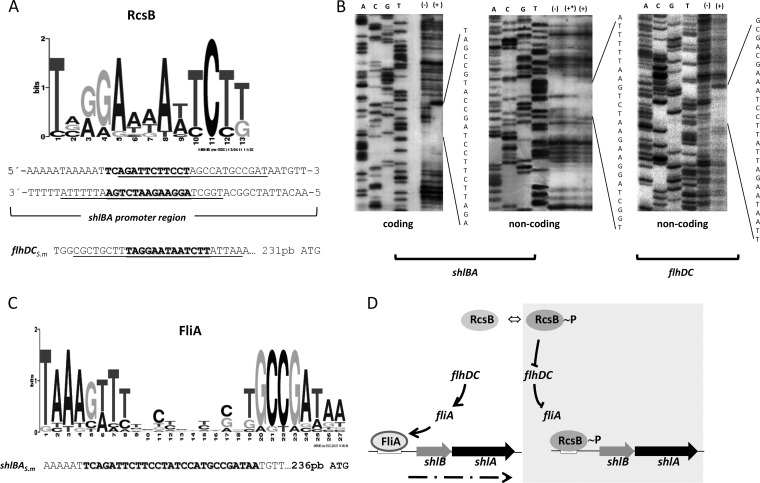
RcsB binds to the promoter region of the S. marcescens shlBA and flhDC operons. (A) Logo showing the consensus motif for the RcsB-binding site and the predicted RcsB-binding site sequences in the promoter regions of S. marcescens (S.m.) flhDC and shlBA. (B) DNA footprinting analysis was performed on end-labeled coding and noncoding strands of the shlB promoter region and on the noncoding strand for the flhDC promoter region. The DNA fragments were incubated with purified RcsB-His×6 at a final concentration of 0 (−) or 400 pmol (+*) or with acetyl-phosphate preincubated with RcsB-His×6 at a final concentration of 400 pmol (+). The positions of the protected areas were determined by comparison with sequence ladders obtained by using the same labeled primer that was used as a probe. The RcsB-protected regions are indicated, with the respective nucleotide sequences. (C) Logo showing the consensus motif for the FliA-binding site and the predicted FliA-binding site sequences in the promoter regions of S. marcescens shlBA. DNA-protected regions obtained by the DNA footprinting assays are underlined (A), while the binding motifs that match the RcsB or FliA consensus motifs are indicated in boldface (C). (D) Schematic overview of the RcsB-dependent regulatory circuit that governs shlBA expression. (Left) When RcsB is inactive (RcsB, nonphosphorylated RcsB protein), FliA protein expression drives shlBA operon transcription by interaction with the FliA-binding motif within the shlBA operon promoter region (dashed arrow, shlBA transcript). (Right) Activated RcsB (RcsB∼P, phosphorylated RcsB protein) represses the transcriptional activity of the flagellar flhDC-dependent transcriptional cascade, inhibiting FliA expression, while it simultaneously downregulates shlBA transcription by direct interaction with the RcsB-binding motif within the shlBA operon promoter region.