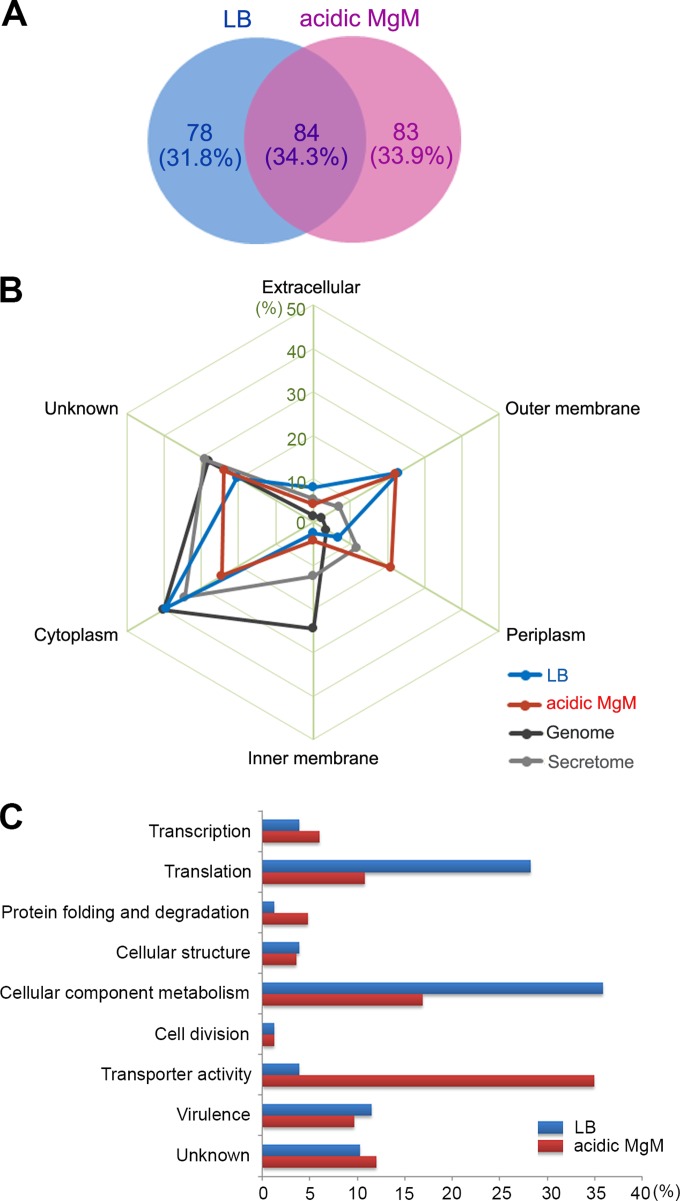
FIG 2.
Proteomic analysis of OMVs isolated under two different growth conditions. (A) Venn diagram of OMV-associated proteins from two proteomic profiles. A total of 245 proteins were identified from the LB-derived and acidic MgM-derived OMVs through LC-MS/MS analysis using the stringent criteria described in Materials and Methods. The proteins found under both conditions or under only one of the conditions were counted, and their numbers (and percentages) are shown. (B) Subcellular location of OMV-associated proteins. The proteins identified through LC-MS/MS profiling were processed using PSORTb (http://www.psort.org/psortb/) to predict their main location in the cell in addition to the OMV. The proteins in the LB-derived and acidic MgM-derived OMV profiles that were predicted to be in the extracellular, outer membrane, periplasm, inner membrane, or cytoplasm fractions were counted, and their percentages are depicted. The proteins identified through S. Typhimurium genome annotation and secretome analysis (see details in Discussion) were subjected to subcellular location prediction and compared with those found in the OMV profiles. (C) Classification of OMV-associated proteins using Gene Ontology (GO)-based functions. The OMV-associated proteins unique to the LB or acidic MgM conditions were grouped based on their biological processes and molecular functions, and their proportions under each condition are plotted.