FIG 1.
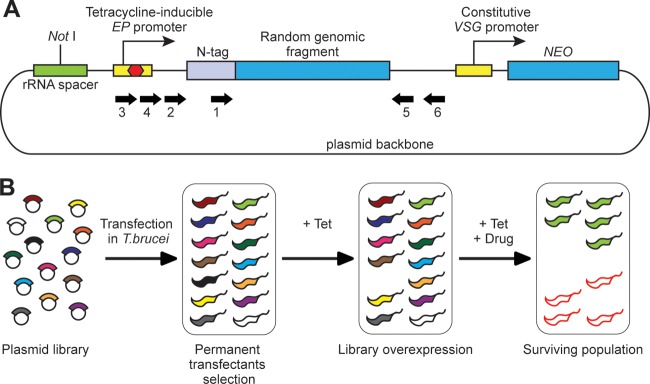
Overexpression approach for drug target discovery in T. brucei. (A) Schematic representation (not in scale) of the overexpression plasmid pHD2542 containing a random genomic fragment. The library insert is expressed with an N-terminal tag, the lambda peptide (N-tag). Expression is controlled by a tetracycline-inducible EP procyclin promoter, which is known to be able to drive overexpression in both bloodstream- and procyclic-form trypanosomes (26). The transfected cells are selected through resistance to neomycin (NEO), stably expressed from a constitutive VSG promoter. The linearized plasmid is integrated into a silent rRNA spacer. The inserts were identified using specific primers (thick arrows). Primer sequences (all from 5′ to 3′) were as follows: primer 1 (illuminaF), TAGCCACCGGGCCC; primer 2 (SeqLib2), CTTGAAGACTTCAATTACACC; primer 3 (SeqLib3), CTATCAGTGATAGAGATCCC; primer 4 (SeqLib4), TTAACATGTTCTCGTCCC; primer 5 (illuminaR), CACACAAATGGATCCTCAGC; primer 6 (LibR), CTGTACGTAAATGTGTTGC. (B) Schematic representation of the approach for drug target screening. The plasmid library is transfected into bloodstream-form T. brucei, and permanent transfectants are selected through resistance to neomycin. Library overexpression is then induced with the addition of tetracycline (+ Tet), and only cells expressing nontoxic polypeptides survive. Following 24-h tetracycline induction, the studied drug is added (+ Drug), and a surviving population is selected after some days. The population is composed of cells overexpressing the target (green with black outlines) and other cells (red outlines) that acquired a growth advantage under drug pressure, presumably through genomic mutations.
