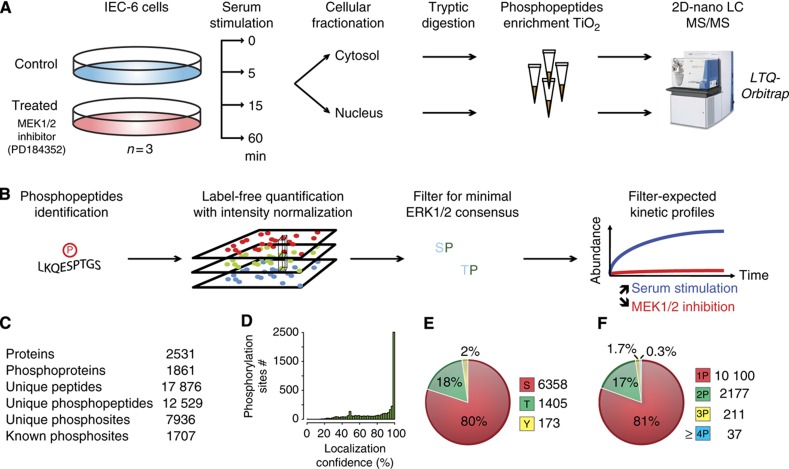
Figure 1.
Experimental workflow and data processing for the identification of candidate ERK1/2 substrates. (A) Experimental workflow for sample processing and MS analysis. (B) Data analysis for the selection of candidate ERK1/2 substrates. (C) Statistics on the number of identified phosphopeptides. (D) Distribution of site-localization confidence data. (E) Distribution of phosphorylated amino acids. (F) Number of phosphorylation sites per peptide.