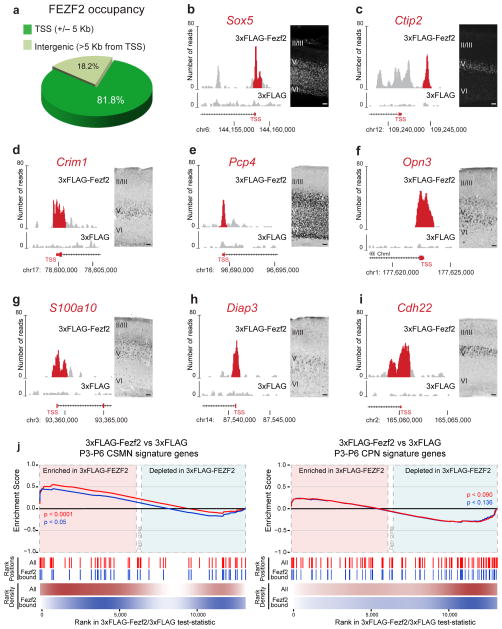
Figure 3.
Genome-wide binding analysis for Fezf2 shows preferential association with proximal promoter regions of CSMN genes. (a) Fezf2 binding events preferentially occur in proximity to promoter regions, within 5 kb of the TSS of annotated genes. (b–i) Examples of 3xFlag-Fezf2 peaks at proximal promoters for early (Sox5 and Ctip2; b,c), middle (Crim1) (d) and late (Pcp4, Opn3, Diap3, S100a10 and Cdh22; e–i) CSMN genes. In situ hybridizations are shown for Crim1 (P21), Pcp4 (P21), Opn3 (P21), S100a10 (P21), Diap3 (P14) and Cdh22 (P21). Immunohistochemistry results are shown for SOX5 (E18.5) and CTIP2 (P1). Scale bars, 100 μm (b–i). (j) The “zero cross” area represents genes where the Cuffdiff2 test statistic is equal to 0 and no appropriate rank information is available. The color scales used for the rank density represent kernel density estimate of gene rank positions (white = 0, red/blue=max density). GSEA for CSMN and CPN signature gene sets. Signature gene sets (in red) and the corresponding subsets bound by Fezf2 (in blue) were assessed for enrichment in Fezf2-overexpressing neurospheres. Both CSMN signature genes and the subset bound by Fezf2 were significantly enriched in Fezf2-overexpressing neurospheres. Neither set of CPN signature genes showed significant enrichment. Source data are shown in Supplementary Table 4.