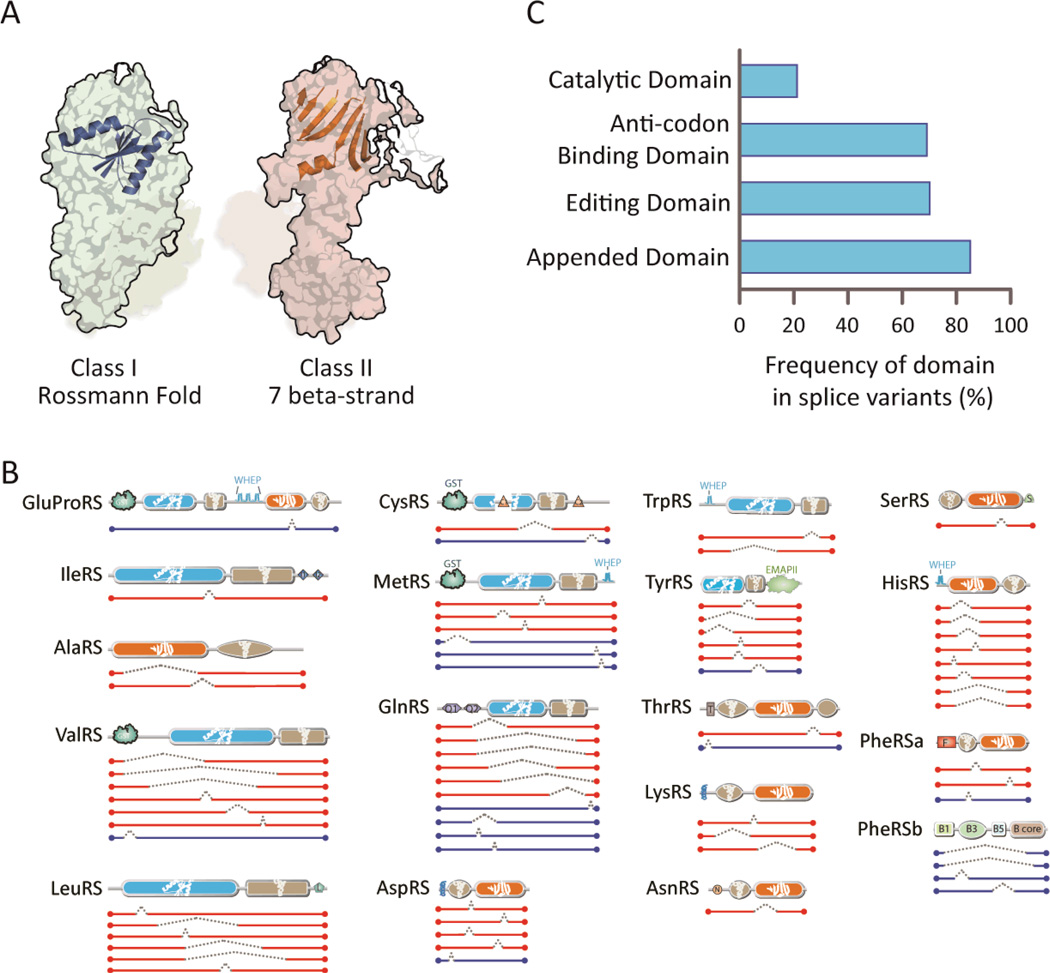
Figure 2. Majority of AARS splice variants are catalytic nulls.
(A) Architectures of the catalytic domains for aminoacylation - Class I versus Class II. The conserved core Rossmann fold is represented on the structure of MetRS (pdb2CT8) (ref: Pubmed 16155581) in Class I, and the conserved core 7 beta-strand with motif-3 helix is represented on the structure of LysRS (pdb4DPG) (ref: Pubmed23159739) in Class II. (B) In-frame splice variants of cytoplasmic AARS are illustrated. Splice variants with catalytic domains deleted (catalytic nulls) are highlighted in red while those with catalytic domains retained are represented in blue. (C) The catalytic domain is abrogated in most AARS splice variants. In contrast, domains that have functions distinct from aminoacylation are predominantly retained. Of note, the UNE domains (such as UNE-S, and UNE-L (abbreviated as “S” and “L”, respectively, in panel B), which comprise part of the “Appended Domain” category and are idiosyncratic to specific synthetases, are retained in the AARS splice variants identified here.