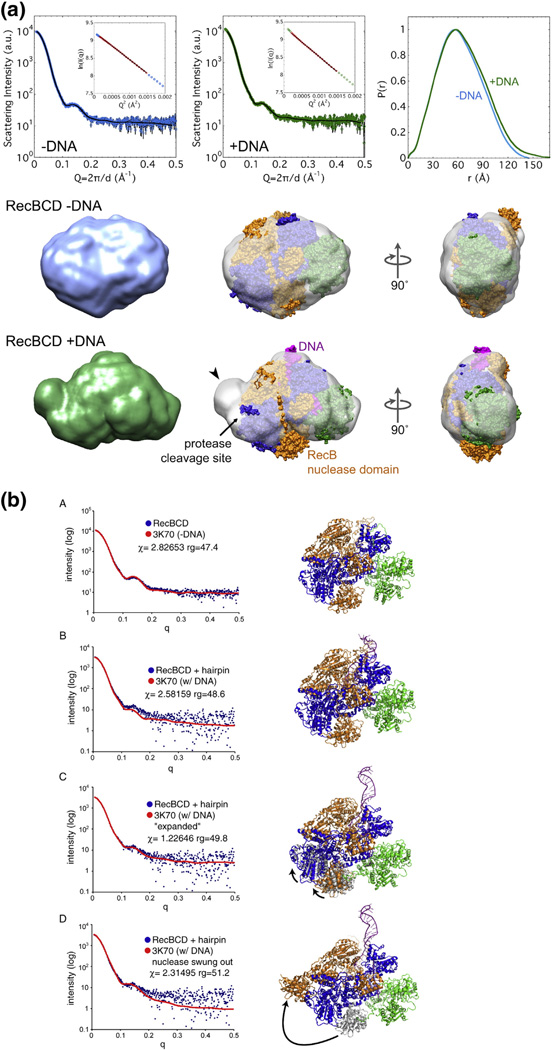
Fig. 9.
SAXS analysis of DNA-bound and unbound RecBCD enzymes. (a) Envelopes of RecBCD enzyme determined by SAXS (data in upper panels) of free enzyme and enzyme bound to hairpin DNA in K-PO4 buffer with glycerol (Figs. S7 and S8; Table S2). Ab initio shape reconstructions agree well with the data; Guinier analyses (insets) show linear fits. The crystal structure, with subunits colored as in Fig. 1a and oriented to be optimally docked within the SAXS envelope, is superimposed on the SAXS envelope. Note that swinging of the RecB nuclease domain (carat) improves the fit and accounts for the change of protease sensitivity (relatively sensitive −DNA and resistant + DNA). (b) SAXS all-atom modeling analysis of the above data. In (b–A), data from enzyme without DNA were tested against PDB structure 3K70 [19] with DNA computationally removed; in (b-B), data from enzyme with a bound DNA hairpin were tested against PDB structure 3K70; in (b–C) and (b–D), data from enzyme with a bound DNA hairpin were tested against two possible conformational changes in the enzyme, one (b–C) involving movement of the lower lobes of RecB and RecC moving toward the helicase domain of RecB, perhaps reflecting jaws closure, and the other (b–D) involving swinging of the nuclease domain to cover the protease-sensitive region. Additional data are in Figs. S7–S16.