Table 7. Comparison of MuGI and JST on simulated and real data, both over 1092 individual sequences of chr1.
| Algorithm | Max. allowed mismatches | RAM usage | |||||
| 0 | 1 | 2 | 3 | 4 | 5 | [GB] | |
| Simulated data | |||||||
| JST-Horspool | 8.0 s | — | — | — | — | — | 2.58 |
| JST-Myers | 22.5 s | 24.0 s | 23.9 s | 24.3 s | 24.5 s | 24.9 s | 2.58 |
MuGI, 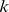 = 30, sparsity = 1 = 30, sparsity = 1 |
8.2 µs | 14.8 µs | 22.7 µs | 33.6 µs | 69.6 µs | 176.0 µs | 1.84 |
MuGI, 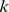 = 30, sparsity = 3 = 30, sparsity = 3 |
10.7 µs | 21.7 µs | 32.4 µs | 47.9 µs | 90.7 µs | 239.0 µs | 0.98 |
| Real data | |||||||
| JST-Horspool | 6.9 s | — | — | — | — | — | 2.58 |
| JST-Myers | 18.4 s | 19.1 s | 19.2 s | 20.0 s | 20.3 s | 20.3 s | 2.58 |
MuGI, 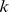 = 30, sparsity = 1 = 30, sparsity = 1 |
7.6 µs | 14.3 µs | 22.1 µs | 53.4 µs | 172.3 µs | 476.5 µs | 1.84 |
MuGI, 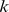 = 30, sparsity = 3 = 30, sparsity = 3 |
12.3 µs | 23.1 µs | 35.3 µs | 72.0 µs | 238.2 µs | 617.9 µs | 0.98 |
MuGI was executed for parameters 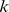 = 30 and sparsities: 1 (index size 2.0 GB), 3 (index size 1.0 GB). Results for JST are averages from only 100 queries due to very long running times. JST times include block generation (blocks of 100K SNPs were used), but in our experiments they are at least an order of magnitude lower than pattern searching. JST-Horspool uses the Boyer–Moore–Horspool exact matching algorithm, while JST-Myers uses Myers' bit-parallel approximate matching algorithm, handling the Levenshtein distance (
= 30 and sparsities: 1 (index size 2.0 GB), 3 (index size 1.0 GB). Results for JST are averages from only 100 queries due to very long running times. JST times include block generation (blocks of 100K SNPs were used), but in our experiments they are at least an order of magnitude lower than pattern searching. JST-Horspool uses the Boyer–Moore–Horspool exact matching algorithm, while JST-Myers uses Myers' bit-parallel approximate matching algorithm, handling the Levenshtein distance (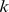 -differences). The JST index size was 468 MB, in addition to the 253 MB of the reference sequence. Note its memory use during the search is significantly higher than the index size and depends on the block size (e.g., its memory use grows to about 13 GB with blocks of 1 M SNPs).
-differences). The JST index size was 468 MB, in addition to the 253 MB of the reference sequence. Note its memory use during the search is significantly higher than the index size and depends on the block size (e.g., its memory use grows to about 13 GB with blocks of 1 M SNPs).
