Figure 1.
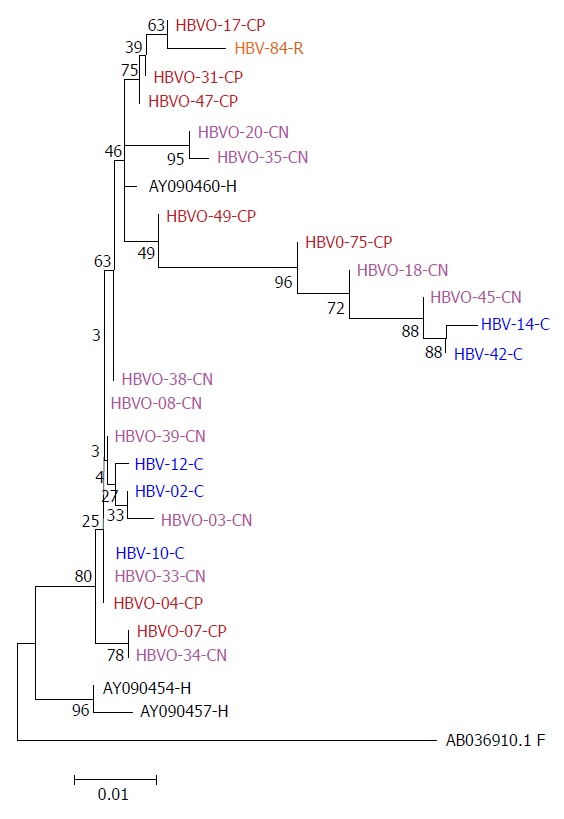
Phylogenetic tree constructed by the Maximum Likelihood method based on the partial nucleotide sequence of the core gen (439 bp) of 18 OHBI isolates (CP and CN); 5 sequences (C) obtained from hepatitis B virus overt co-infection (HbsAg+), using the three genotype H reference sequences from Genbank (AB375159.1, AB375160.1, AB375161.1), as a reference group and one hepatitis B virus sequence from a chronic hepatitis B virus mono-infected patient (HBV-84-R). Genetic distances were estimated using the Kimura two-parameter matrix and Bootstrap values are indicated for the major nodes as a percentage of the data obtained from 1000 replicates. C: Core; -CN: Seronegative OHBI; -CP: Seropositive OHBI; -S: HBsAg+ isolates.
