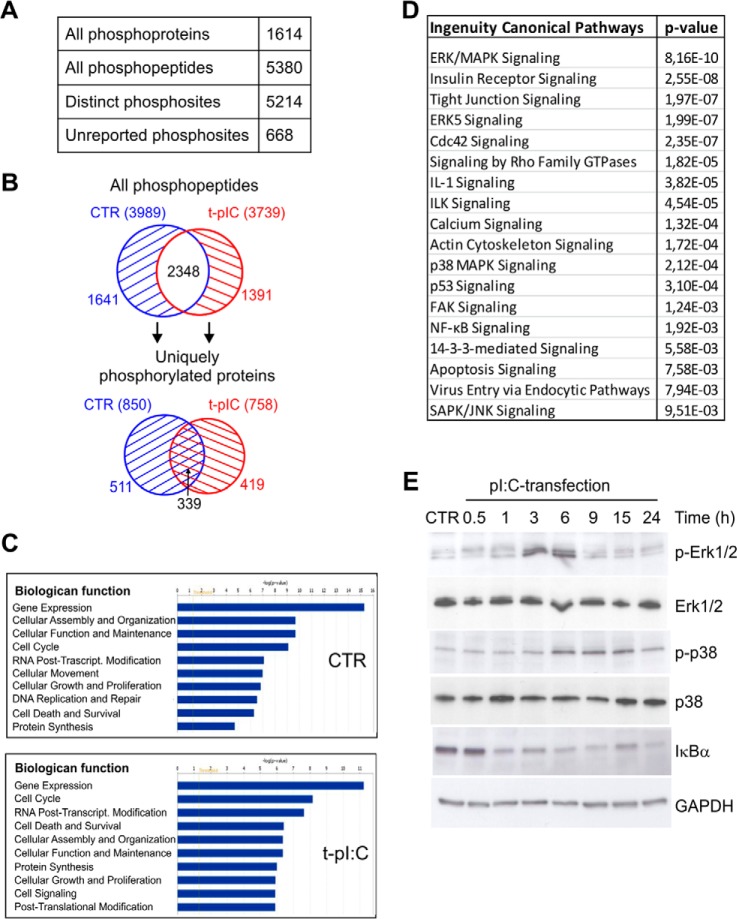
Fig. 2.
Phosphoproteome analysis of dsRNA-stimulated human keratinocytes. A, identified phosphoproteins and phosphopeptides and distinct phosphorylation sites. B, the distributions of all identified phosphopeptides (upper panel) and uniquely phosphorylated proteins (lower panel) in control and dsRNA-stimulated samples. C, most significant biological functions of uniquely phosphorylated proteins in control (850) and dsRNA-induced (758) samples were determined using IPA. D, selected canonical pathways activated during viral dsRNA stimulation (complete list in supplemental Table S2). IPA analysis was done with the 419 uniquely phosphorylated proteins identified only after pI:C transfection. Highly similar canonical pathways were identified when the phosphoproteins identified in at least two out of three biological replicates were taken into analysis (supplemental Table S2). E, HaCaT keratinocytes were transfected with pI:C for the indicated times, after which cell lysates were subjected to SDS-PAGE and immunoblotting using antibodies against phosphorylated p38 MAPK (Thr180/Tyr182), total p38, phosphorylated Erk1/2 (Thr202/Tyr204), total Erk1/2, or IκBα. GAPDH detection was used to confirm the equal loading.