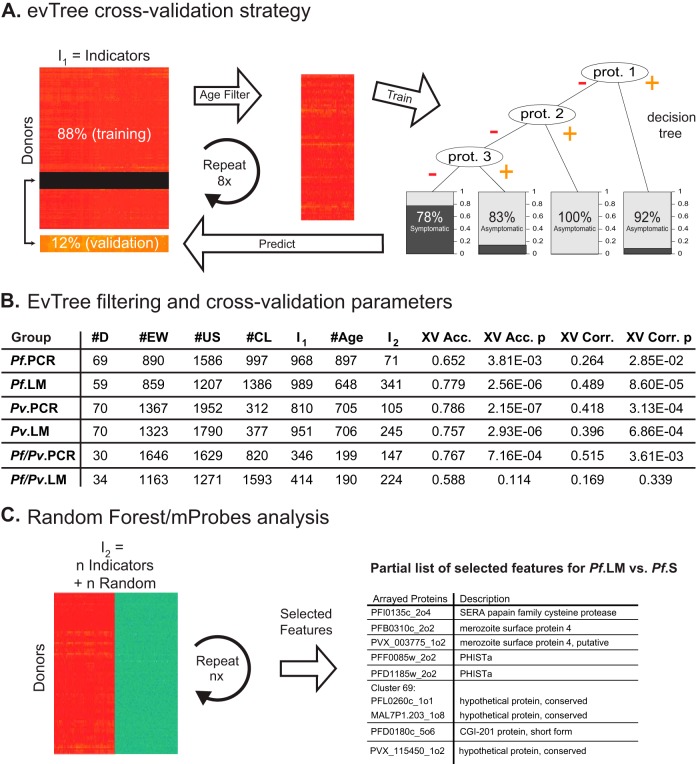
Fig. 3.
Machine-learning approach for biomarker selection. A, evTree cross-validation strategy used to compare antibody responses in each of the six asymptomatic groups to those in the corresponding symptomatic groups. After dividing each dataset into an 88% training set and a 12% validation set and applying the age filter, the evTree algorithm was trained with 8-fold cross-validation to generate decision trees that predicted whether donors were symptomatic or asymptomatic based on their antibody responses to subsets of arrayed proteins. B, Results of filtering and evTree cross-validation parameters for each of the six pairwise comparisons. The table shows the number of donors in each comparison (#D); the number of responses removed by the empty well (#EW), U.S. naïve donor (#US), and clustering (#CL) filters; the number of responses that remained after these filters were applied (I1); the number of responses removed by the age filter (#Age) and the number of responses that remained after it was applied (I2); the cross-validated accuracy (XV Acc.) and p value (XV Acc. p) of the resulting classifier; as well as the corresponding Matthew's correlation coefficient (XV Corr.) and p value (XV Corr. p) for the classifier. C, Overview of mProbes with Random Forest feature selection strategy used to identify antibody responses that discriminate between symptomatic and asymptomatic donors. After adding noise by randomizing labels for indicators that remain rafter the Age filter was applied (I2), the algorithm identifies features that distinguish between symptomatic and asymptomatic donors. Shown is a representative subset of features selected from the Pf. LM versus Pf.S pairwise comparison.