Abstract
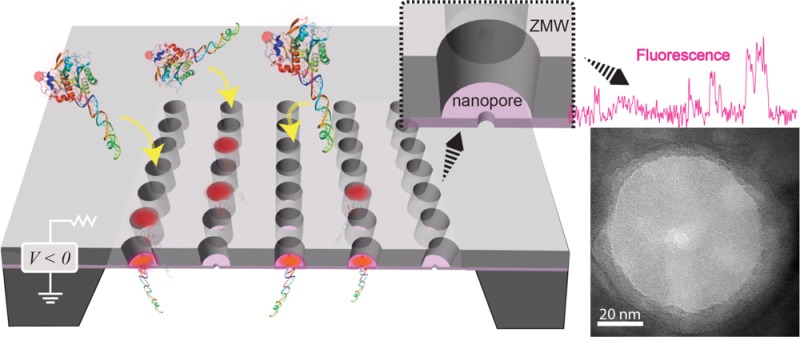
We have developed a hybrid nanopore/zero-mode waveguide device for single-molecule fluorescence and DNA sequencing applications. The device is a freestanding solid-state membrane with sub-5 nm nanopores that reversibly delivers individual biomolecules to the base of 70 nm diameter waveguides for interrogation. Rapid and reversible molecular loading is achieved by controlling the voltage across the device. Using this device we demonstrate protein and DNA loading with efficiency that is orders of magnitude higher than diffusion-based molecular loading.
Keywords: SMRT-sequencing, DNA sequencing, single molecule, nanophotonics, zeptoliter
The ability to isolate and study the dynamics of individual biomolecules using fluorescence has revolutionized our understanding of basic mechanisms in biology. Single-molecule fluorescence relies on the detection of photon emission from individual labeled molecules, which is often complicated by various factors including interference from neighboring molecules, a limited fluorescence lifetime due to photobleaching, and background optical noise from other molecules in the bulk solution. Zero-mode waveguides (ZMWs), nanostructures that comprise subwavelength cylindrical wells in an opaque metallic film,1 alleviate most of the challenges of single-molecule fluorescence. When illuminated, an exponentially decaying electric field forms at the ZMW base. Combined with the subwavelength lateral field confinement of the aperture ZMWs can achieve zeptoliter excitation volumes.2 This confined illumination has been exploited for single molecule measurements of lipid diffusion in a bilayer,3,4 reverse transcription,5 DNA methylation,6 and translation,7 among other biophysical processes.8 Notably, the ZMW is an essential element of the Single Molecule, Real-Time (SMRT) DNA sequencing platform, where single DNA polymerases inside the ZMWs incorporate phosphate-labeled, color-coded nucleotides.9
A key requirement for ZMW-based studies is that a single biomolecular entity occupies each ZMW. In SMRT sequencing, the yield of singly occupied ZMWs determines the overall efficiency, accuracy, and cost of DNA sequencing. Immobilization of single polymerases inside ZMWs is currently achieved using either diffusion or magnetic beads. Both modes of loading result in a theoretical maximum of 37% of singly occupied ZMWs due to a Poissonian statistical limit.10 In addition to single molecular occupation inside ZMWs, the kinetics and efficiency of DNA template loading also suffer in diffusion-based approaches, which can be a major hurdle for sequencing applications in which only low DNA concentrations are available, e.g., epigenetic analysis of mammalian cellular DNA.6 Diffusion-based DNA loading for SMRT sequencing typically requires 30–60 min exposure at ∼80 pM DNA concentrations, and input DNA requirements further increase for fragments longer than 1000 base pairs.11 This has limited most epigenetic SMRT sequencing applications to bacterial studies,6,12,13 or created the necessity to develop chemically sophisticated enrichment methods for applicability to mammalian DNA samples.14 For magnetic-bead-based loading, the efficiency of immobilizing long DNA molecules is increased, translating to ∼3–30 pM input concentrations, although hour-long immobilization times are needed.
The need for single occupation and time-efficient molecular loading for ZMW-based studies are both compromised by the reliance on diffusion- or bead-assisted loading. While a recent report that employs DNA origami scaffolds has allowed super-Poissonian occupation of single biotin groups at ZMW bases,15 the authors note an occupation limit even using a high origami concentration, as well as a significant fraction of doubly occupied ZMWs. In contrast to diffusion, active methods that focus and manipulate molecules in space have the potential to greatly enhance ZMW-based studies. The ability to actively draw single molecules from bulk and position them inside the illumination volumes of ZMWs would impact DNA sequencing and a wide array of immobilization-based ZMW applications by enhancing the efficiency, sensitivity, and accuracy of the devices. Vice versa, reversing the conditions under which molecules are focused inside the ZMWs can release a used molecular complex from the ZMW.
Solid-state nanopores have the ability to focus and trap single molecules at specific, nanometer-precise positions. These pores are milled using electron or ion beams16−18 to form nanometer-scale holes through synthetic insulating membranes. Applying a transmembrane voltage across an electrolyte-immersed pore generates a steady-state ion current through the nanopore constriction, resulting in a highly localized electric field profile in the pore vicinity that can be used to focus and capture individual charged biomolecules.19 These pores have been suggested for nanopore-based DNA sequencing technology20 and for delivering biomolecules to a variety of patterned nanostructures.21−27 Recent experiments have used large (>50 nm) metallized nanopores to detect metal nanoparticles28 and fluorescently labeled DNA,29 although no mechanism is provided in this work for immobilizing and precisely positioning them at a desired position within the ZMW.
In this article, we demonstrate ZMW devices equipped with nanopore-based biomolecular positioners at their base. Specifically, we have developed an integrated nanopore–ZMW device (NZMW) in which a ZMW array is fabricated on top of an ultrathin silicon nitride (SiNx) membrane, such that sub-5 nm diameter nanopores are present at the base of ZMWs on the membrane. Using these devices we demonstrate the controlled immobilization/ejection of DNA–protein complexes from the pore, as well as a vastly improved loading kinetics of large DNA molecules (6000 base pairs) into the ZMWs over diffusion-based loading.
Device Characterization
A schematic illustration of our NZMW single-molecule positioners is shown in Figure 1a. The device consists of a silicon chip that has been processed using established methods2 to contain an array of ZMWs on a ∼100 × 100 μm2 freestanding SiNx membrane (see Supporting Information for details). The ZMW arrays were passivated from reaction with piranha solution by depositing an 11 nm-thick SiO2 layer using atomic-layer deposition.30 Nanopores were then drilled in predetermined locations in the ZMW arrays using transmission electron microscopy (TEM), followed by treatment with piranha solution in order to hydrate the pores prior to experiments (see Supporting Information). The device was assembled in a custom cell that allows fluidic access to both sides of the membrane, as well as optical access to the bottom side of the chip as shown in Figure 1a (see Supporting Information).
Figure 1.
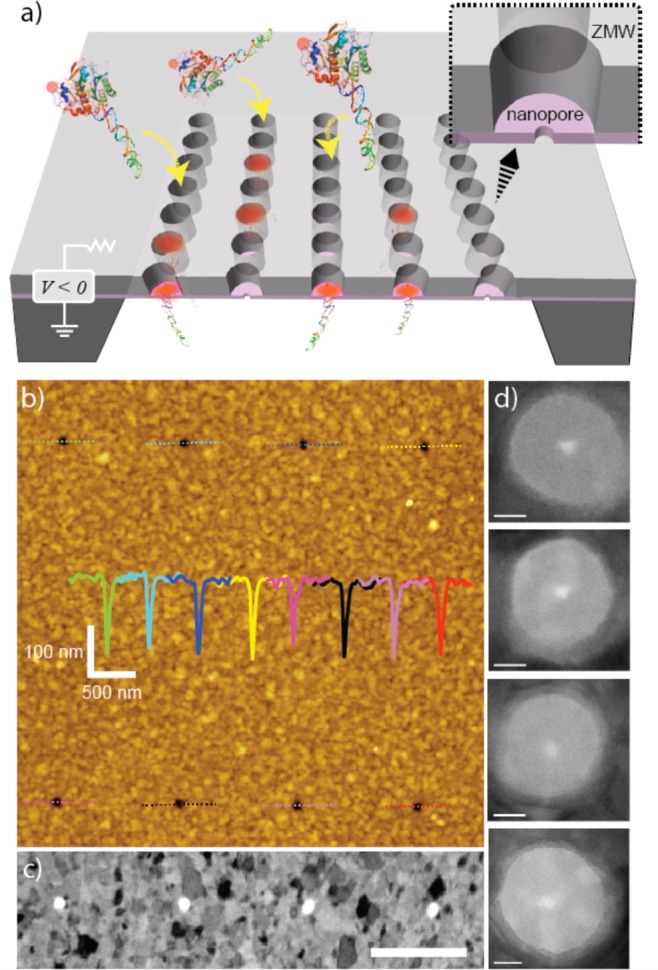
NZMW device. (a) Scheme of the NZMW. An array of ZMWs is positioned on a 35 nm silicon nitride membrane with nanopores at the bases of waveguides (inset). A voltage bias actively draws complexes of biotinylated DNA and fluorescently labeled streptavidin to the pore, which places the fluorophore in the ZMW excitation volume. (b) An AFM scan of the ZMW membrane illustrates the topography of the surface. ZMWs are spaced 1.3 μm × 4 μm. Line scans of each ZMW demonstrate uniformity with an average top diameter of 86.2 ± 6.4 nm (N = 21). The scans have a pointed bottom profile because the AFM tip cannot penetrate the full depth of the waveguide. (c) Dark-field scanning transmission electron micrograph (inverted contrast) of four ZMWs in the array (scale bar = 1 μm). (d) TEM images of ZMWs with 3 to 3.5 nm nanopores drilled in their centers (scale bars = 20 nm). ZMWs have a measured base diameter of 64.9 ± 3.7 nm (N = 57).
Figure 1b shows an AFM scan of a 2 × 4 ZMW array on a SiNx membrane (4 μm × 1.3 μm spacing). The ZMWs are seen as dark uniform circles in the image for which height profiles through the ZMW centers are shown as insets to the figure. Although the base of the ZMWs cannot be accessed in AFM due to the tip geometry, the height profiles reveal uniform top diameters measured to be 86.2 ± 6.4 nm (N = 21). The base diameters were measured by dark-field scanning TEM (Figure 1c) and bright-field TEM (Figure 1d) to be 64.9 ± 3.7 nm (N = 57). The dark-field images of Figure 1c in which the contrast was inverted for clarity, display a polycrystalline structure with grains in the range of 50–150 nm, characteristic of a thermally evaporated metal film. The images in Figure 1d show four typical NZMWs that contain 3–3.5 nm diameter pores drilled at their center. We note that the TEM images shown in Figure 1 represent the first noncross-sectional view (e.g., top-view) of ZMWs using the TEM, because prior ZMW devices have all been fabricated on ∼100 μm thick glass substrates that are too thick for TEM imaging. On the basis of these AFM and TEM measurements of the top and base diameters, respectively, we arrive at a funnel-like ZMW shape, as previously obtained with other ZMW devices fabricated on fused silica substrates.2
Capture of charged molecules into the NZMWs can be greatly impacted by an electric field gradient present near the NZMW volume. It is established that DNA capture into a nanopore is strongly assisted by the residual electric field near the pore mouth,19 which generates a localized electromotive force that migrates the molecule and focuses it to the pore. To examine the impact of ZMW presence on the electric field profile near the pore, we used finite-element simulations to numerically compute the voltage profile in the vicinity of a 3 nm diameter nanopore in the absence (Figure 2a) and presence (Figure 2b) of a 60 nm diameter ZMW above it. As the simulations show, the addition of the ZMW constriction results in an electric field gradient with significant presence beyond the ZMW top. The dotted contour lines, which indicate the positions at which the voltage drop is one percent of the total trans-membrane bias, highlight a 4-fold extension of the field away from the pore. This extended field facilitates the migration of charged biomolecules toward the ZMW volume.
Figure 2.
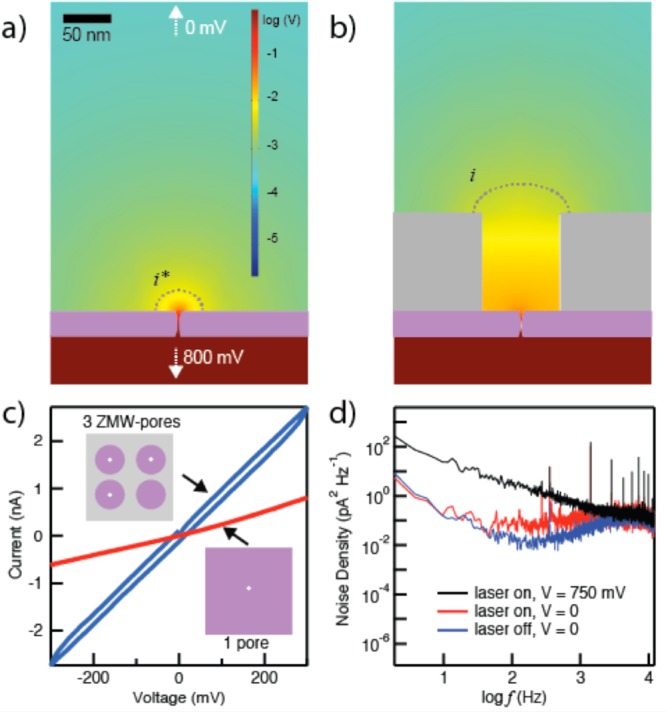
Electrical properties of the NZMW. Numerical solution for the voltage profile induced by applying 800 mV to (a) a 3 nm pore without a ZMW and (b) a 60 nm diameter ZMW with a 3 nm pore ([KCl] = 400 mM, T = 25 °C). Dotted lines i* and i indicate the equipotential contour line where the voltage drop is 1% of the total transmembrane voltage. (c) I–V curve for an array of three NZMWs in 400 mM KCl (blue curve) compared to that for a SiNx pore under the same conditions (red curve). (d) Power spectral density of electrical noise for a NZMW membrane under different experimental conditions as indicated in the legend (λ = 488 nm, P = 20 mW, ∼40 W/cm2 sample intensity).
Figure 2c plots an I–V curve measured on an array of three NZMWs with 3 nm diameter pores in 400 mM KCl (blue) as well as an I–V curve of a single 3 nm diameter pore (red). In both cases, the I–V curves are linear, indicating open pores are present in the devices. The minor hysteresis in the I–V curve of the NZMW array is an artifact of the additional capacitance of the ZMW structure, which does not adversely interfere with our ability to capture and observe molecules inside NZMWs. Power spectral densities (PSD) of the current noise for an NZMW device under various experimental conditions are shown in Figure 2d. The PSD plots show a typical shape for nanopore measurements, characterized by 1/f noise at low frequencies, thermal (Johnson) noise at intermediate frequencies, and capacitive-dominated noise at high frequencies.31−33 Laser illumination at zero bias (red curve) affects the thermal noise, while having little impact on the 1/f and capacitive regimes. In contrast, upon application of voltage (black) the 1/f noise dominates the PSD, as previously observed in nanopore experiments.34 Despite the presence of ZMWs on the membrane, the overall noise is comparable to that of conventional SiNx nanopores.32
Detecting DNA/Protein Complexes
Using a device that contains a single NZMW with a 2.5 nm diameter nanopore, we demonstrate the ability to capture a DNA/protein complex and dissociate its biotin–streptavidin bond in a ZMW under high bias. A solution that contains 1003 bp 5′-biotinyated DNA complexed to Alexa Fluor 647-labeled streptavidin (see Materials and Methods) was added to the cis chamber, which resulted in voltage-driven electrophoretic focusing of the complexes into the ZMW volume. When the DNA threads into the pore, the force on the DNA against the streptavidin that is anchored to the ZMW base causes the eventual dissociation of the complex.35 Mounting our custom cell on an inverted microscope equipped with 640 nm laser illumination (Coherent Cube, Coherent, Inc.) and emCCD detection (see Supporting Information) we simultaneously recorded nanopore current and NZMW fluorescence. Upon application of 850 mV, a stable open pore baseline current was observed followed by a stochastic series of spikes that correspond to DNA and/or DNA/streptavidin interactions with the nanopore in the NZMW. In addition, we observed occasional long-lived events (>1 s) that correspond to long-lived presence of the complex within the nanopore. These long-lived events were coincident with discrete increases in fluorescence from the NZMW (Figure 3, points 1–5). Notably, in events 1 and 2 of Figure 3 we observed relatively shallow current blockades, which may represent a complex present in the NZMW without one of its DNA molecules being fully threaded. This explanation is supported by events 1 and 2, which respectively show a complex temporarily adhering before diffusing away, and a complex remaining near the pore during which we observed other DNA translocation events. For events 3–5, we observed deeper blockade levels accompanied by increases in fluorescence, which indicate full DNA threading and streptavidin presence at the NZMW base. Dissociation of the biotin–streptavidin bond at high voltage has previously been observed in a solid-state nanopore under similar applied voltage values.35 In each of these events, the simultaneous reduction of the nanopore current and increase in fluorescence indicates the capture of individual DNA/protein complexes in the pore. Finally, reversal of the voltage results in immediate ejection of the complex from the NZMW (e.g., events 2, 4, and 5), as observed by a coincident decrease in fluorescence intensity.
Figure 3.

Reversible positioning of a single DNA–protein complex inside a NZMW. Simultaneous current (250 kHz sampling, 10 kHz filtering) and fluorescence (1 pixel region of interest, 10.02 ms exposure time, signal-averaged to 400 ms) traces from a single NZMW containing a 2.5 nm pore for a sample of biotinylated 1003-bp DNA conjugated to Alexa Fluor 647-labeled streptavidin. Brief translocation spikes are translocations of free DNA. Points 1–5 identify events where a fluorescently labeled DNA–protein complex entered the ZMW illumination volume and occluded the pore, resulting in simultaneous fluorescence from the NZMW and blockage of the nanopore current. The inset schematically depicts the experimental scheme. DNA is pulled into the pore but prevented from translocating by the streptavidin, giving long-lasting current blockage. While immobilized in the pore, the labeled streptavidin sits in the ZMW excitation volume, resulting in fluorescence.
Simultaneous Positioning of DNA–Protein Complexes inside NZMWs
ZMW devices are ideal for high-throughput fluorescence-based biomolecular analysis, which requires immobilization of the molecule inside the ZMW excitation volume for extended periods of time. We have tested the principle of voltage-driven capture of multiple complexes in a 2 × 4 array of NZMWs that contain 3–4 nm diameter nanopores, as shown in Figure 4. We imaged the membrane while applying alternating biases of +500 and −500 mV to trap and eject the same DNA/streptavidin complex as used for the experiment in Figure 3. Figure 4a shows a fluorescence image of the NZMW array (left), as well as a series of three images during different time periods of the experiment. The bright spots in the images represent fluorescence that is due to occupied NZMWs. We note that five of the eight NZMWs in the array were active during the experiment, with the remaining NZMWs not displaying optical signal. This yield of ∼60% is a reasonable yield of active nanopores in this diameter range. In Figure 4b we plot time traces of fluorescence from five NZMWs, identified as 1–5 in Figure 4a, as well as from a ZMW that does not contain a nanopore, labeled as “N”. At the beginning of the trace a (+) voltage was applied, during which molecules are clearly observed in the ZMW volume. With the exception of pore 1 (indicated by *), application of (−) voltage resulted in ejection of complexes from the NZMWs, as indicated by a return of the fluorescence signal to the baseline level. Upon restoring the (+) voltage we observed fluorescence activity in all five NZMWs, indicating molecular loading. This infrequent occurrence of noncorrelated signals in NZMW 1 is a possible result of protein sticking to the surface of the device.36
Figure 4.
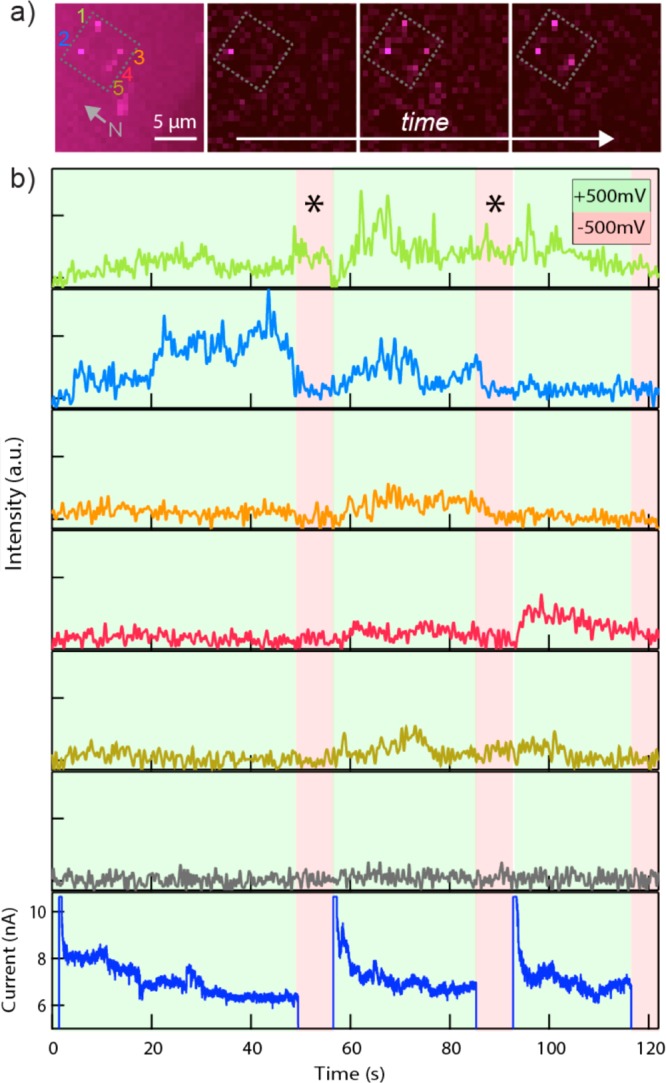
Immobilization of DNA–protein complexes in an array of NZMWs. (a) Fluorescence images of a 2 × 4 NZMW array (enclosed by dotted line) with immobilized complexes of 1003 bp biotinylated DNA and Alexa Fluor 647-labeled streptavidin. Five NZMWs that captured molecules are identified in the leftmost image, which is a projection of all frames from the experiment. The point identified N is a nearby ZMW with no pore. The next three images from different points in the experiment illustrate molecules entering and leaving NZMWs. (b) Fluorescence traces from ZMWs 1–5 and N (1 pixel region of interest for each NZMW, 42.55 ms exposure time, signal-averaged to 500 ms) superimposed with membrane bias. In regions with green background, the transmembrane voltage is 500 mV. In regions with red background, it is −500 mV. * identifies points where a protein adheres to the membrane, resulting in fluorescence persisting through negative voltage pulses.
We note that while activity was seen in many of the NZMW devices, no activity was observed in the remaining ZMWs that contain no nanopores (e.g., ZMW “N” in Figure 4). We suggest two main reasons for this observation: First, the radius of gyration of a 1003 bp DNA molecule is 40 nm, which is slightly larger than the ZMW radius (35 nm). This mismatch presents an energy barrier for diffusion of the DNA–streptavidin complex into the ZMW. Second, because we have not applied chemistry to covalently link the diffusing DNA to the ZMW surface, there is no mechanism to immobilize the complex in the ZMWs.
Focusing Long DNA Fragments into ZMWs
Finally, we investigate the efficiency of DNA capture into NZMW devices. A solution of 230 pM 6000 bp DNA labeled with YOYO-1 intercalating dye (10:1 bp/dye ratio, 488 nm excitation, see Materials and Methods) was placed on the cis side of the membrane. To monitor DNA entry, we imaged a ZMW array that contained a single NZMW while the applied voltage was toggled between +850 and −850 mV. Figure 5 shows fluorescence traces from the NZMW, as well as traces from three representative ZMWs. The inset shows three fluorescence images of the device that correspond to a time-averaged stack of frames from the whole experiment (i), as well as time-integrated images at negative (ii) and positive (iii) voltages. The NZMW (red arrow) was clearly visible based on its notable fluorescence at positive voltage values, while the remaining three ZMWs did not exhibit a voltage-induced fluorescence enhancement (see Supporting Information). Similarly, the traces in Figure 5 clearly show distinct entry of individual DNA molecules into the NZMW volume, as indicated by a stochastic set of fluorescence enhancement spikes. We find DNA capture to be highly efficient; the on-time of DNA within the NZMW was 51% when the voltage was (+), whereas the off-time was >99% for negative voltages (see Supporting Information). Additionally, we find a prolonged 6.0 ± 5.5 s mean duration of fluorescence spikes, during which we observe a very dynamic fluorescence signal that points to stochastic DNA fluctuations within the ZMW that occur on a slow time scale.
Figure 5.
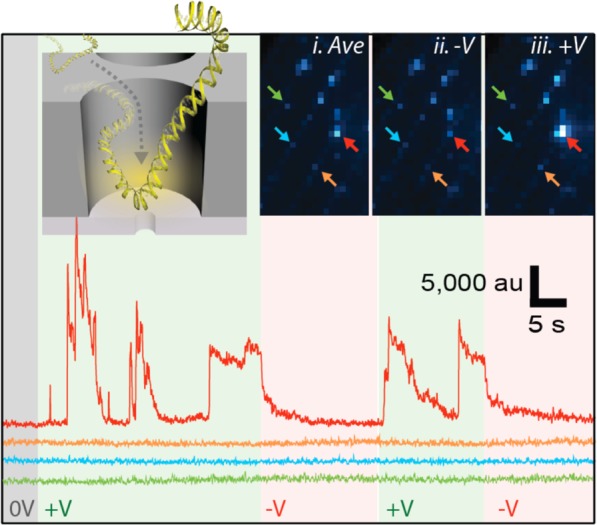
DNA focusing into a NZMW. Fluorescence time traces from a single NZMW that contains a 3 nm diameter pore in an array of ZMWs is monitored for the fluorescence from 6000 bp DNA labeled with YOYO-1 (9 pixel region of interest for each NZMW, 10.8 ms exposure time, signal-averaged to 100 ms). Inset illustrates DNA entering the illumination volume of a NZMW as it migrates toward the pore, resulting in increased ZMW fluorescence. ZMW arrays are shown in fluorescence images (i)–(iii) with (i) being an averaged image of all frames in the experiment, and (ii) and (iii) being the membrane under respective −850 and 850 mV. Colored arrows identify ZMWs with corresponding colored fluorescence traces in bottom plot. The red arrow identifies a NZMW. Green and red backgrounds in the fluorescence traces correspond to periods of positive and negative voltage, respectively (see Supporting Information for electrical trace).
To quantify the DNA loading we compared the on-time of the NZMW with on-times of other neighboring ZMWs in our experiment for times in which positive voltage was applied. For the random sample of 13 ZMWs, we have analyzed the resulting ratio of on-times tNZMW/tZMW is 580, highlighting the utility of nanopores as biomolecular focusing elements for ZMW-based studies (see Supporting Information). From a SMRT-sequencing perspective, we also compare the input DNA requirements for NZMWs to those of ordinary ZMWs. A typical protocol for diffusive loading of a 2000 bp template uses a 150 pM DNA concentration and 60 min of reaction time, yielding a concentration-normalized loading rate of 1.9 × 10–6 pM–1 s–1. Magnetic bead loading results in improvements on the concentration requirement (3–30 pM) but still requires long incubation times (60 min) for optimal Poisson loading, translating to a concentration-normalized loading rate of 1.7 × 10–5 pM–1 s–1. In contrast, based on the mean DNA arrival time in our NZMW experiment (3.5 s), the loading rate in NZMWs is 1.3 × 10–3 pM–1 s–1, orders-of-magnitude more efficient than in the case of diffusive or magnetic bead loading.
We have demonstrated a novel device that consists of nanopores at the base of ZMWs for efficient and versatile positioning of single molecules. The fabrication process for these devices involved a combination of electron-beam and photolithography methods and resulted in the first demonstration of ZMWs on freestanding SiNx membranes that contain nanopores at their bases. Using synchronous optical and electrical recordings, we have demonstrated the reversible, voltage-driven positioning and ejection of individual DNA–protein complexes, as well as a mechanism for greatly enhancing the entry of long DNA molecules into ZMWs. We note that loading of long DNA molecules into ZMWs for SMRT sequencing is inefficient because of the large DNA coil size with respect to the ZMW dimensions. This need to “package” DNA into ZMWs results in a conformationally restricted DNA that is unlikely to encounter a DNA polymerase at the ZMW base on short time scales.37 Current protocols for activating ZMWs for sequencing involve incubation of the ZMWs with a preformed complex of DNA and a streptavidin-polymerase fusion protein, which still results in a slow binding to the ZMW surface due to the imposed conformational restriction.11 The need to prereact DNA and polymerase in solution, as well as the need for higher DNA concentrations, has limited certain studies involving precious DNA samples using this method. Finally, we have demonstrated the first ZMW platform in which the ZMW can be reused by releasing a molecular complex from the ZMW volume at the click of a button. The combination of ZMWs and nanopores greatly increases the efficiency of DNA loading, which can aid in the development of future SMRT sequencing applications in genetics and epigenetics. In addition, this ability to focus, hold, and release biomolecules from the illumination volume of the ZMW should allow many biophysical studies at the molecular level.
Materials and Methods
Sample Molecule Preparation
DNA–protein complexes were prepared from PCR-synthesized biotinylated DNA and Alexa Fluor 647-labeled streptavidin (Life Technologies, Carlsbad, CA). Biotinylated DNA was incubated with labeled streptavidin at a 4:1 DNA/streptavidin ratio for 15 min (see gel image in Supporting Information). YOYO-labeled DNA was prepared from 6000 bp DNA (Thermo Scientific, Tewksbury, MA) and YOYO-1 intercalating dye (Life Technologies, Carlsbad, CA). DNA and dye were incubated for 20 min at 50 °C with a 10:1 base pair/dye molar ratio.
Numerical Simulations
Voltage distributions near pores (Figure 2a,b) were computed with COMSOL Multiphysics (COMSOL, Burlington, MA). The Poisson–Nernst–Planck equations were numerically solved for a geometry consisting of two micron-scale cylindrical compartments (i.e., cis and trans) connected by a nanopore embedded in a perfectly insulating membrane. An element size as fine as 0.1 nm and additional boundary meshing layers inside the pore were used to ensure no edge effects skew the physical results. A positive bias voltage of 800 mV was enforced at the bottom surface of the trans chamber and ground to the top surface of the cis chamber.
Data Acquisition and Analysis
AFM scans were taken with a Bruker FastScan AFM in tapping mode. TEM imaging and pore fabrication were performed with a JEOL 2010FEG (Northeastern University). NZMW chips were cleaned for 5 min in heated piranha solution, rinsed thoroughly in deionized water, dried under vacuum, and immediately assembled for experiment in a PEEK flow cell (see Supporting Information). The cell was mounted in a Faraday cage on the stage of an Olympus IX81 inverted microscope with a 60×, 1.2 NA water immersion objective. The membrane was illuminated with a Coherent Cube 640 nm laser and a Coherent Sapphire 488 nm laser. An Axopatch 200B amplifier was used for current monitoring off Ag/AgCl electrodes. Electrical data was recorded using custom-made LabVIEW software (National Instruments, Woburn, MA). Images were taken with a Hamammatsu ImagEM EMCCD, recorded with HCImage Live software (Hamamatsu, Sewickley, PA), and analyzed with ImageJ.
Acknowledgments
We acknowledge Alan Bleier, Meredith Metzler, and Rob Ilic of the Cornell NanoScale Facility (CNF); Stephen Jones for aid in fabrication; Wentao Liang for aid in TEM imaging; Spencer Carson for the COMSOL simulations; Andrey Ivankin for the optical cell design. This work was supported by NIH award no. R21-HG006873 (M.W. and J.K.). This work was performed in part at the CNF, a member of the National Nanotechnology Infrastructure Network (NNIN), which is supported by the National Science Foundation (Grant ECCS-0335765), and at the Center for Nanoscale Systems (CNS), a member of the NNIN, which is supported by the National Science Foundation under NSF award no. ECS-0335765. CNS is part of Harvard University.
Supporting Information Available
Supporting Information contains details on device fabrication, device preparation, experimental apparatus, DNA/streptavidin complex characterization, and data analysis. This material is available free of charge via the Internet at http://pubs.acs.org.
M.F., S.W.T., and J.K. are full-time employees at Pacific Biosciences, a company developing sequencing technologies.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Levene M. J.; Korlach J.; Turner S. W.; Foquet M.; Craighead H. G.; Webb W. W. Science 2003, 299, 682–686. [DOI] [PubMed] [Google Scholar]
- Foquet M.; Samiee K. T.; Kong X. X.; Chauduri B. P.; Lundquist P. M.; Turner S. W.; Freudenthal J.; Roitman D. B. J. Appl. Phys. 2008, 103, 034301. [Google Scholar]
- Edel J. B.; Wu M.; Baird B.; Craighead H. G. Biophys. J. 2005, 88, L43–L45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samiee K. T.; Moran-Mirabal J. M.; Cheung Y. K.; Craighead H. G. Biophys. J. 2006, 90, 3288–3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilfan I. D.; Tsai Y. C.; Clark T. A.; Wegener J.; Dai Q.; Yi C. Q.; Pan T.; Turner S. W.; Korlach J. J. Nanobiotechnol. 2013, 11, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flusberg B. A.; Webster D. R.; Lee J. H.; Travers K. J.; Olivares E. C.; Clark T. A.; Korlach J.; Turner S. W. Nat. Methods 2010, 7, 461–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura S.; Aitken C. E.; Korlach J.; Flusberg B. A.; Turner S. W.; Puglisi J. D. Nature 2010, 464, 1012–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P.; Craighead H. G. Annu. Rev. Biophys. 2012, 41, 269–293. [DOI] [PubMed] [Google Scholar]
- Eid J.; Fehr A.; Gray J.; Luong K.; Lyle J.; Otto G.; Peluso P.; Rank D.; Baybayan P.; Bettman B.; Bibillo A.; Bjornson K.; Chaudhuri B.; Christians F.; Cicero R.; Clark S.; Dalal R.; Dewinter A.; Dixon J.; Foquet M.; Gaertner A.; Hardenbol P.; Heiner C.; Hester K.; Holden D.; Kearns G.; Kong X. X.; Kuse R.; Lacroix Y.; Lin S.; Lundquist P.; Ma C. C.; Marks P.; Maxham M.; Murphy D.; Park I.; Pham T.; Phillips M.; Roy J.; Sebra R.; Shen G.; Sorenson J.; Tomaney A.; Travers K.; Trulson M.; Vieceli J.; Wegener J.; Wu D.; Yang A.; Zaccarin D.; Zhao P.; Zhong F.; Korlach J.; Turner S. Science 2009, 323, 133–138. [DOI] [PubMed] [Google Scholar]
- Korlach J.; Marks P. J.; Cicero R. L.; Gray J. J.; Murphy D. L.; Roitman D. B.; Pham T. T.; Otto G. A.; Foquet M.; Turner S. W. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 1176–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacific Biosciences Template Preparation and Sequencing Guide; Pacific Biosciences of California: Menlo Park, CA, 2014. [Google Scholar]
- Clark T. A.; Murray I. A.; Morgan R. D.; Kislyuk A. O.; Spittle K. E.; Boitano M.; Fomenkov A.; Roberts R. J.; Korlach J. Nucleic Acids Res. 2012, 40, e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G.; Munera D.; Friedman D. I.; Mandlik A.; Chao M. C.; Banerjee O.; Feng Z.; Losic B.; Mahajan M. C.; Jabado O. J.; Deikus G.; Clark T. A.; Luong K.; Murray I. A.; Davis B. M.; Keren-Paz A.; Chess A.; Roberts R. J.; Korlach J.; Turner S. W.; Kumar V.; Waldor M. K.; Schadt E. E. Nat. Biotechnol. 2012, 30, 1232–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C. X.; Clark T. A.; Lu X. Y.; Kislyuk A.; Dai Q.; Turner S. W.; He C.; Korlach J. Nat. Methods 2012, 9, 75–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pibiri E.; Holzmeister P.; Lalkens B.; Acuna G. P.; Tinnefeld P. Nano Lett. 2014, 14, 3499–3503. [DOI] [PubMed] [Google Scholar]
- Li J.; Stein D.; McMullan C.; Branton D.; Aziz M. J.; Golovchenko J. A. Nature 2001, 412, 166–169. [DOI] [PubMed] [Google Scholar]
- Storm A. J.; Chen J. H.; Ling X. S.; Zandbergen H. W.; Dekker C. Nat. Mater. 2003, 2, 537–540. [DOI] [PubMed] [Google Scholar]
- Yang J. J.; Ferranti D. C.; Stern L. A.; Sanford C. A.; Huang J.; Ren Z.; Qin L. C.; Hall A. R. Nanotechnology 2011, 22, 285310. [DOI] [PubMed] [Google Scholar]
- Wanunu M.; Morrison W.; Rabin Y.; Grosberg A. Y.; Meller A. Nat. Nanotechnol. 2010, 5, 160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally B.; Singer A.; Yu Z.; Sun Y.; Weng Z.; Meller A. Nano Lett. 2010, 10, 2237–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A. P.; Instuli E.; McGilvery C. M.; Baldwin G.; McComb D. W.; Albrecht T.; Edel J. B. Nano Lett. 2010, 11, 279–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshiro T.; Matsubara K.; Tsutsui M.; Furuhashi M.; Taniguchi M.; Kawai T. Sci. Rep. 2012, 2, 501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson M. P.; Dekker C. Nano Lett. 2013, 13, 1029–1033. [DOI] [PubMed] [Google Scholar]
- Puster M.; Rodríguez-Manzo J. A.; Balan A.; Drndić M. ACS Nano 2013, 7, 11283–11289. [DOI] [PubMed] [Google Scholar]
- Traversi F.; Raillon C.; Benameur S. M.; Liu K.; Khlybov S.; Tosun M.; Krasnozhon D.; Kis A.; Radenovic A. Nat. Nanotechnol. 2013, 8, 939–945. [DOI] [PubMed] [Google Scholar]
- Ivanov A. P.; Freedman K. J.; Kim M. J.; Albrecht T.; Edel J. B. ACS Nano 2014, 8, 1940–1948. [DOI] [PubMed] [Google Scholar]
- Zhao Y.; Ashcroft B.; Zhang P.; Liu H.; Sen S.; Song W.; Im J.; Gyarfas B.; Manna S.; Biswas S.; Borges C.; Lindsay S. Nat. Nanotechnol. 2014, 9, 466–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini M. P.; Wiener A.; Turek V. A.; Chon H.; Lee S.; Ivanov A. P.; McComb D. W.; Choo J.; Albrecht T.; Maier S. A.; Edel J. B. Nano Lett. 2013, 13, 4602–4609. [DOI] [PubMed] [Google Scholar]
- Auger T.; Mathé J.; Viasnoff V.; Charron G.; Di Meglio J.-M.; Auvray L.; Montel F. Phys. Rev. Lett. 2014, 113, 028302. [DOI] [PubMed] [Google Scholar]
- Hausmann D.; Becker J.; Wang S. L.; Gordon R. G. Science 2002, 298, 402–406. [DOI] [PubMed] [Google Scholar]
- Smeets R. M. M.; Keyser U. F.; Dekker N. H.; Dekker C. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabard-Cossa V.; Trivedi D.; Wiggin M.; Jetha N. N.; Marziali A. Nanotechnology 2007, 18, 305505. [Google Scholar]
- Rosenstein J. K.; Wanunu M.; Merchant C. A.; Drndic M.; Shepard K. L. Nat. Methods 2012, 9, 487–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P.; Mitsui T.; Farmer D. B.; Golovchenko J.; Gordon R. G.; Branton D. Nano Lett. 2004, 4, 1333–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabard-Cossa V.; Wiggin M.; Trivedi D.; Jetha N. N.; Dwyer J. R.; Marziali A. ACS Nano 2009, 3, 3009–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedzwiecki D. J.; Grazul J.; Movileanu L. J. Am. Chem. Soc. 2010, 132, 10816–10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemla Y. R.; Ha T. Science 2014, 345, 380–381. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


