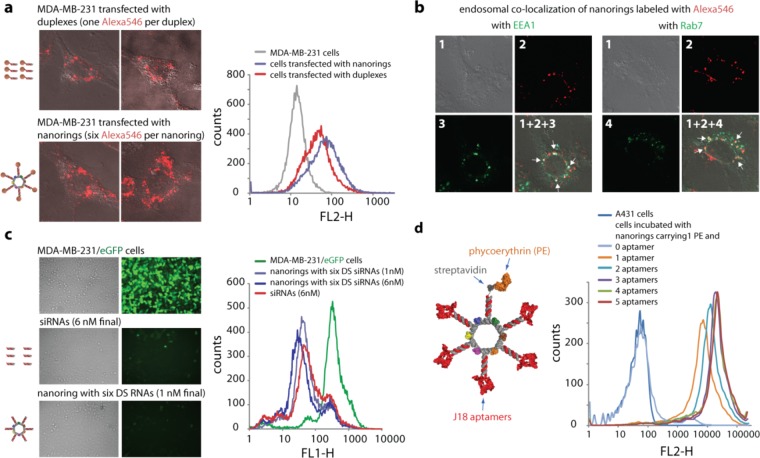
Figure 3.
Cell uptake, endosomal colocalization, silencing, and RNA aptamer mediated binding efficiencies of functional nanorings. (a) Transfection efficiencies using human breast cancer cells (MDA-MB-231). DS RNAs (60 nM final) covalently labeled with one Alexa 546 per duplex were compared to the functionalized nanorings (10 nM final) labeled with six Alexa 546 dyes. One day after the transfection, the efficiencies were analyzed by confocal fluorescence microscopy and flow cytometry experiments. (b) Studying the localization of nanorings with commonly used markers for endosomal compartments Early Endosome Antigen 1 (EEA1) and Rab7. (c) GFP knockdown assays using human breast cancer cells (MDA-MB-231/GFP) which stably express enhanced GFP (eGFP). Fluorescence microscopy (left panel) and statistical analysis (30000 cells per sample) of flow cytometry experiments (right panel) of eGFP expression 3 days after the transfection of cells with DS RNA duplexes and nanorings functionalized with six DS RNAs against eGFP. The ratio of DS RNA duplexes to DS RNA functionalized nanorings was 6:1. (d) Nanorings labeled with phycoerythrin (PE) and containing different numbers of the EGFR-specific J18 aptamer selected to specifically bind EGFR expressed on A431 cells were tested for relative binding efficiencies using FACS. The J18 RNA aptamer model is a conceptual cartoon, based on the minimum free energy secondary structure (MFE). Image numbers in (b) correspond to differential interference contrast (DIC) images (1), Alexa546 emission (2), EAA1 antibody staining (3), and Rab7 antibody staining (4). Images (1 + 2 + 3) and (1 + 2 + 4) are superpositions of three different images.