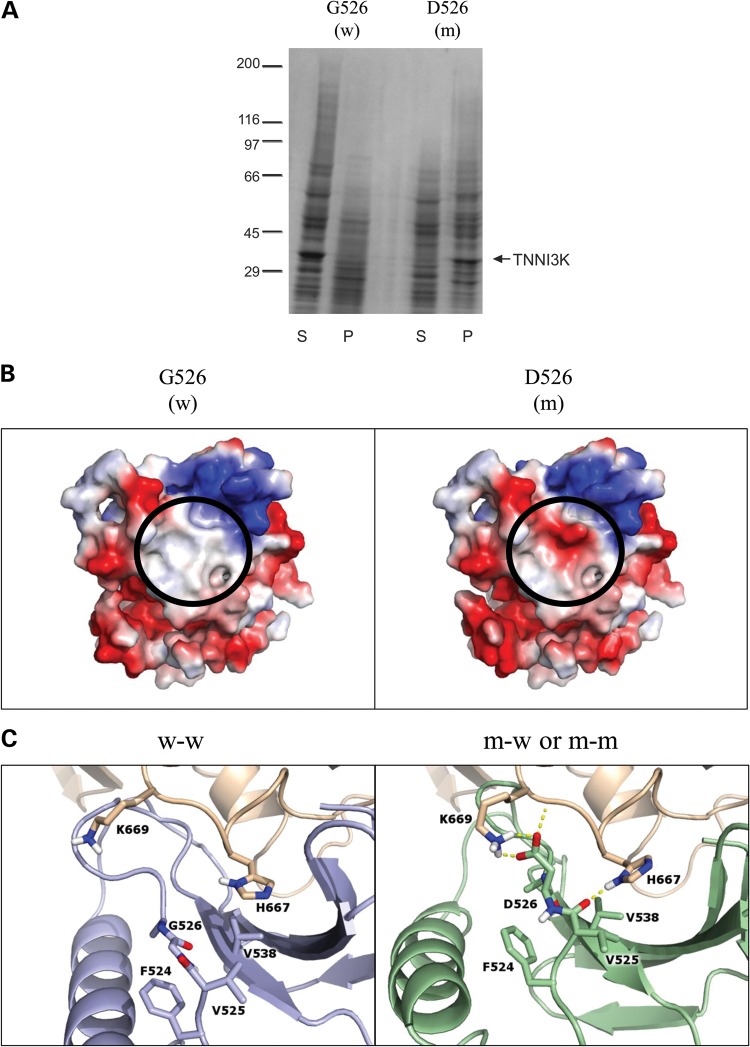
Figure 4.
In vitro expression and in silico modeling of mutant peptide. (A) Kinase domains of wild-type (w; G526) and mutant (m; D526) TNNI3K were expressed and the resulting pellet (P) and supernatant (S) fractions were analyzed by SDS-Page and Coomassie Blue, demonstrating poor solubility of the mutant peptide. The arrow indicates the 32 kilodalton kinase domain of TNNI3K protein and the molecular weight standards (×10−3) are shown on the left. (B) Electrostatic protein modeling (21) highlights a large hydrophobic patch on the surface of the kinase domain (white), which becomes negatively charged (red) in the presence of the G526D mutation. (C) The left panel depicts docking of wild-type (blue) to another wild-type TNNI3K (tan) (w–w) whereas the right panel shows the docking pose for mutant TNNI3K (green) to either wild-type or mutant TNNI3K (both represented in tan) (m-w and m–m). Residues within 3.5 Å of residue 526 in either pose are shown in atomic detail. Hydrogen bonds involving these indicated residues are drawn as dashed yellow lines. When mutated, the loop containing residue 526 shifts toward the binding partner to make more contacts including an electrostatic lysine-aspartic acid interaction (depicted as h-bonds).