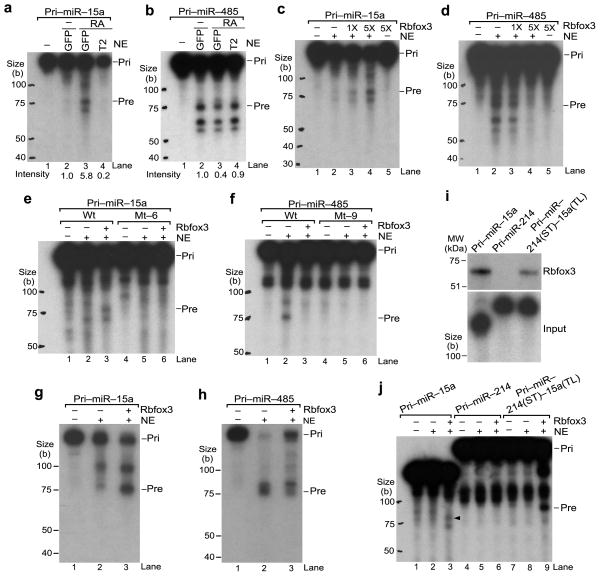
Figure 7.
Direct effects of Rbfox3 on pri-miRNA processing into pre-miRNA. (a, b) In vitro pri-miRNA processing by NE from different conditions of P19 cell cultures. Samples are RNA products from radiolabeled pri-miRNA which was incubated with NE or without NE (−) from RA-treated P19-GFP, P19-T2 cells or untreated (−) P19-GFP cells. Autoradiograms following urea-PAGE are shown. Original pri-miRNA (pri) and processed pre-miRNA (pre) are indicated. Intensity numbers under panels indicate relative pre-miRNA abundance compared to that in untreated P19-GFP. (c, d) In vitro pri-miRNA processing by NE with Rbfox3. Samples are RNA products from radiolabeled pri-miRNAs incubated in NE from untreated P19-GFP cells with increasing amounts (1X, 5X) of in vitro synthesized myc-Rbfox3 or with control reticulocyte lysates (myc-Rbfox3,−). (e, f) In vitro processing of mutant pri-miRNAs. Mt-6 and mt-9 are the same as those in Figure 6. (g, h) RNA blot analysis of in vitro processed RNAs from pri-miRNAs. Samples are unlabeled pri-miRNA incubated with (+) or without (−) NE and myc-Rbfox3 as indicated. RNA blots using radiolabeled oligoDNAs complementary to the mature miRNA sequences are shown. (i) In vitro UV crosslinking of Rbfox3 with the chimeric pri-miRNA containing the terminal loop of pri-miR-15a. Upper panel shows an autoradiogram of myc-Rbfox3 crosslinked with radiolabeled pri-miRNAs following LDS-PAGE. Lower panel shows input pri-miRNAs following Urea-PAGE. (j) In vitro processing of the chimeric pri-miRNA containing the pri-miR-15a terminal loop in the presence of Rbfox3. An arrowhead indicates pre-miR-15a. Uncropped image of panels i is provided in Supplementary Figure 5.