Figure 2.
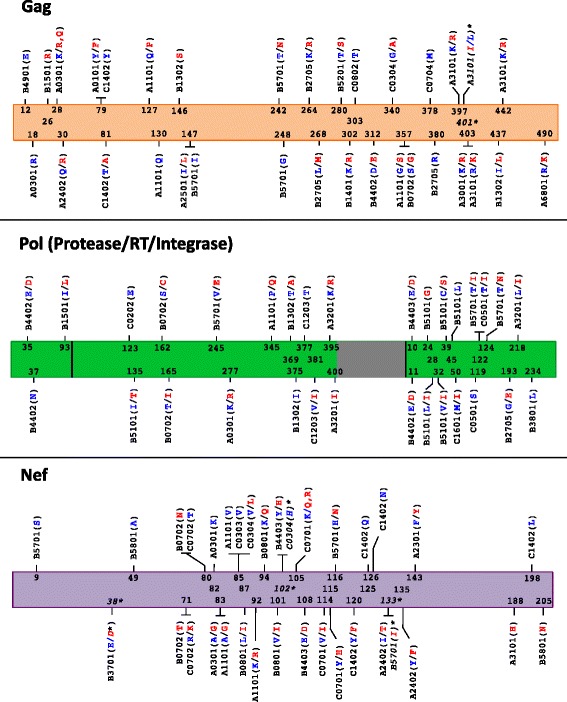
HLA-associated polymorphisms detectable at the population level in cohorts of the present size and genetic composition. Gag, Pol and Nef “immune escape maps” indicate the codon location, specific amino acid residues and HLA restrictions of the N = 162 known HLA-associated polymorphisms detectable at the population level in cohorts of the present size and host/viral genetic composition. “Nonadapted” amino acids (those under-represented in the presence of the HLA allele) are blue; these represent the “immunologically susceptible” form for the HLA allele in question (and usually represent the subtype consensus residue). “Adapted” amino acids (those over-represented in the presence of the HLA allele) are red; these represent the HLA-associated “escape variant”. Adapted and Nonadapted associations are counted independently; in most cases both forms are detectable at the population level at a given p-value threshold (e.g. at Gag codon 242, “T” and “N” represent the B*57:01 – associated nonadapted and adapted forms, respectively), whereas in other cases only one of the two forms is detectable at a given threshold (e.g. at Gag codon 12, “E” represents the B*49:01-associated nonadapted form but no specific adapted form is detected at this threshold). Asterisks (*) and italicized text denote the five HLA-associations at Gag codons 401 and Nef codons 38, 102 and 133 that were defined via detection in the early cohort at p < 0.01, but were p ≥ 0.01 in the chronic cohort. Note that HIV-1 RT genotyping was performed for codons 1–400 of this protein only (the remainder of RT is colored gray). Subsequent analyses focused on this list of HLA-associated polymorphisms.
