Figure 4.
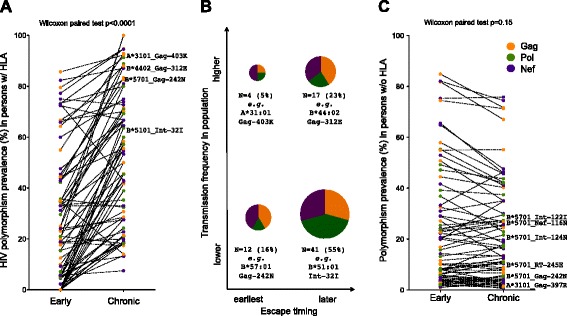
Estimated extent of escape and reversion between early and chronic infection. Panel A: For each of the specific HLA-associated “adapted” (escaped form) HIV-1 polymorphisms investigated (N = 74 total), the proportion of persons expressing the restricting HLA and harboring the relevant polymorphism (“proportion escaped”) in early versus chronic infection are depicted as linked pairs. The data indicate that, on average, escape prevalence in persons expressing the restricting HLA allele more than doubles between early and chronic infection. Panel B: The N = 74 HLA-associated “adapted” polymorphisms are broken down in terms of their relative timeline of escape (“earlier” vs. “later”, where the former is defined as population-level signal of p < 0.05 in early infection) as well as their relative prevalence/transmission frequency in the population (“lower” vs. “higher”, where the former is defined as <30%). The size of each pie reflects the proportion of polymorphisms in each category, while the “pie slices” denote the breakdown of polymorphisms by HIV-1 protein (orange, green and purple for Gag, Pol and Nef respectively). Summary statistics and examples of HLA-associated polymorphisms in each category are also provided. Panel C: The proportion of persons harboring an HLA-associated adapted HIV-1 polymorphism in the absence of the restricting HLA allele in early versus chronic infection is shown. The data are generally consistent with slow reversion of many transmitted escape mutations [7,14,34]. In all panels, polymorphisms are colored by HIV-1 protein: Gag (orange), Pol (green) and Nef (purple); those mentioned in the text are labeled.
