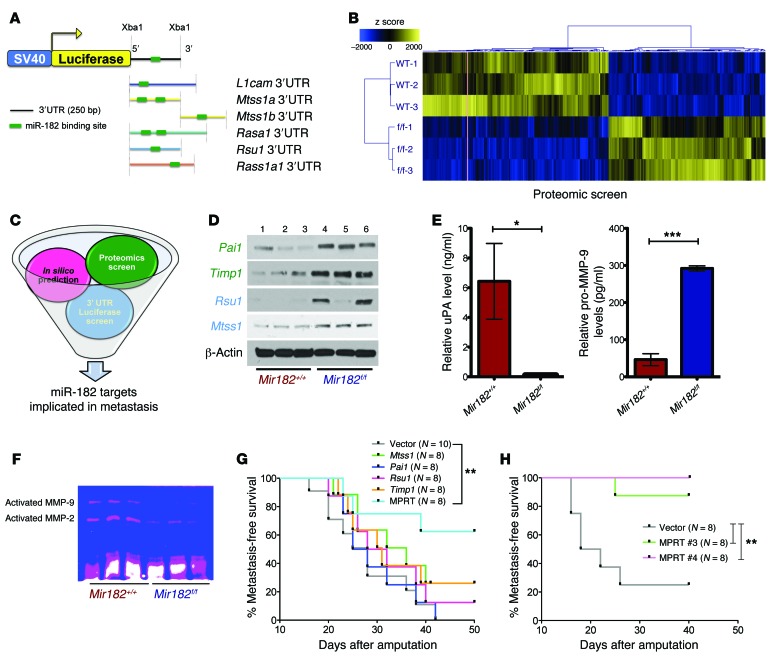
Figure 6. miR-182 target genes cooperate to regulate metastasis.
(A) In silico/3′ UTR luciferase screen used to predict miR-182 targets (top panel) and number of putative miR-182–binding sites on respective 3′ UTRs of the different genes. (B) Heat map shows differential expression of approximately 400 proteins between miR-182 WT and miR-182 deleted primary sarcoma cells from a proteomic screen. (C) Schematic shows the different approaches used to identify miR-182 target genes implicated in metastasis. (D) Western blot validates upregulation of Pai1, Timp1, Rsu1, and Mtss1 in miR-182–deleted KP cells (1–3, different KPY Mir182 WT; 4–6, different KPY Mir182f/f cell lines). (E) ELISA validates decreased uPA and increased unprocessed pro–MMP-9 in the cell-culture media from miR-182–deleted primary sarcoma cells. (F) Zymography confirms decreased activation of MMP-2 and MMP-9 in the cell-culture media from miR-182–deleted KP cells. (G and H) Kaplan-Meier curves show that ectopic expression of all 4 miR-182 targets together (H), but not individual targets alone (G) are required to rescue miR-182–mediated metastasis in an orthotopic metastasis assay. Mice were euthanized 40 days following amputation. Five mice had microscopic lung nodules despite having no clinical evidence of metastasis (Supplemental Figure 13G). MPRT, cell line expressing all 4 miR-182 targets, i.e., Mtss1, Pai1, Rsu1, and Timp1. Two-tailed Student’s t test and 1-way ANOVA were used for statistical analysis. All data are mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.005.