Figure 1.
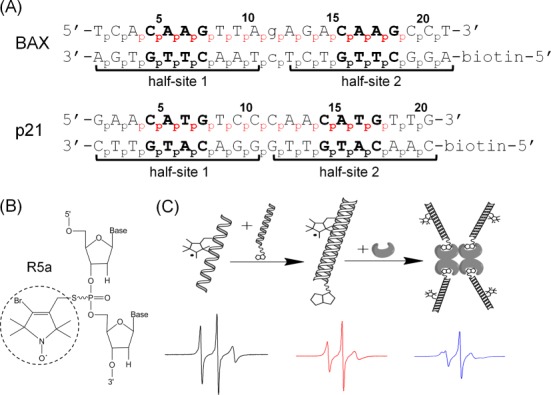
Experimental design. (A) Sequences of DNA duplexes studied. Both duplexes are REs of the p53 tumor suppressor, with two decameric half-sites (marked by brackets) each containing a ‘CWWG’ core (bold). Lower case letters in the BAX duplex mark the one-base-pair spacer between the two half-sites. No spacer is present in p21. For each duplex, the numbering scheme of the phosphates is shown, and the spin-labeled sites are indicated in red. (B) Chemical structure of the R5a spin label. (C) A schematic showing EPR sample assembly. One of the DNA strands was spin-labeled (left), paired with the complementary biotinylated strand (middle), then tethered to a streptavidin tetramer to form an ∼103 kD complex (right). Representative EPR spectra are shown below each step.
