Figure 2.
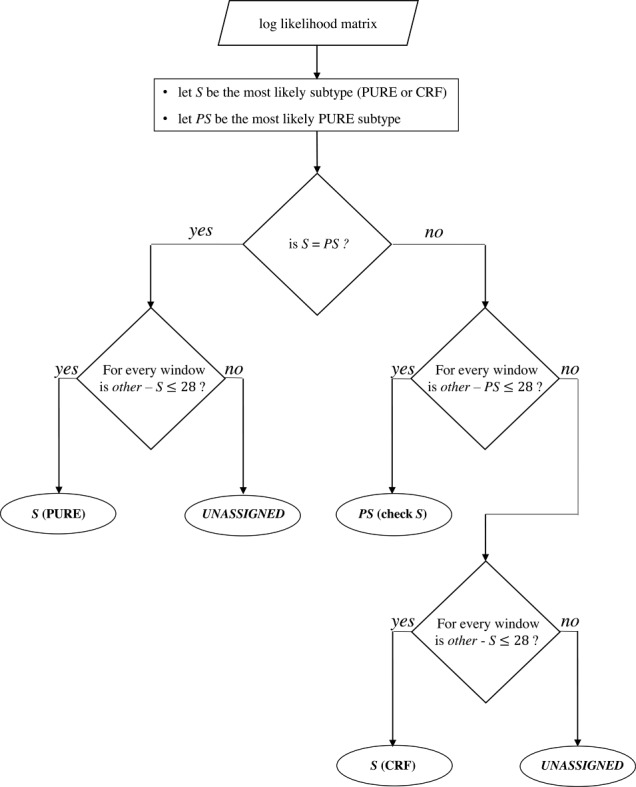
Subtype decision tree. The row sums of the log-likelihood matrix provide the overall likelihood of the query sequence to belong to each subtype. These sums are ordered to identify the most likely subtype (S) and the most likely pure subtype (PS). If the query sequence has the highest likelihood of belonging to a pure subtype (i.e. S = PS), this likelihood is challenged against the likelihoods of the sequence to be of any other subtype (other, PURE or CRF) by sliding over the matrix by 100-bp windows with a stepping size of 3 bp. If the difference between the row sums within the current window remains below the recombination threshold (i.e. 28) for each window, the pure subtype is assigned. Otherwise, COMET returns the result ‘UNASSIGNED’. If the query sequence has the highest likelihood of being a CRF, COMET performs a similar challenge, but only against the most likely pure subtype (PS) at first. If this difference remains below the recombination threshold (i.e. 28), COMET assigns the pure subtype (S) with an indication to check for the CRF, indicating a region where the CRF is pure. If the difference is higher than the recombination threshold, a second scan is performed as for the PURE situation, challenging each subtype against the initially assigned CRF.
