Figure 10.
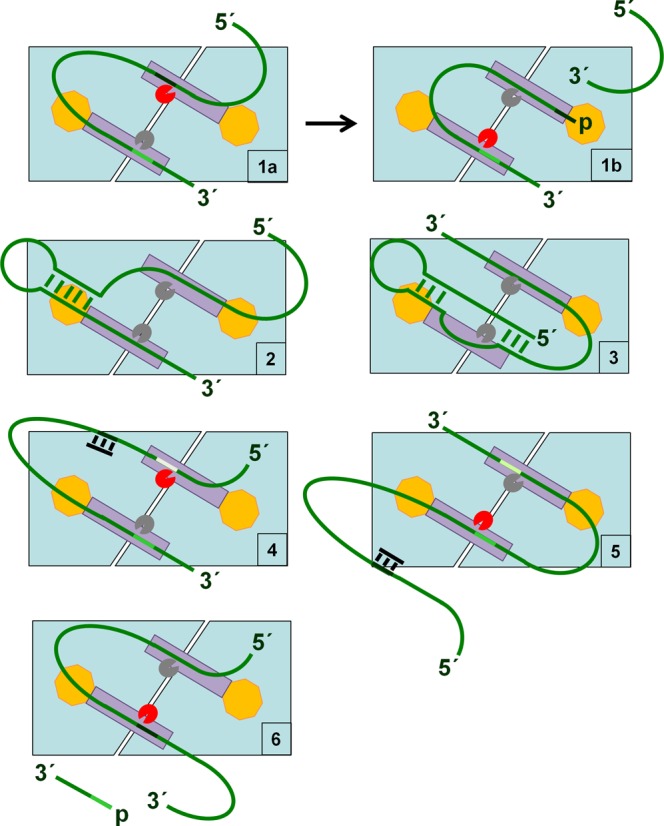
Direct entry: a flexible mode of RNase E cleavage. The principal dimer of RNase E is shown schematically (pale blue). It can interact simultaneously with RNA (green strand) via two unpaired regions (handholds) using its two equivalent RNA-binding channels (dark grey rectangles) (panel 1a). Whilst grasping two unpaired regions, one can be cleaved. The active sites are indicated by pacman symbols, which are coloured red when preferential cleavage of the corresponding unpaired regions is described. Cleavage of the 5′ most site (panel 1a) produces a 5′-monophosphorylated end that, following repositioning within the channel, provides a foothold for the 5′ sensor domain (yellow octagon), thereby extending the overall interaction to facilitate cleavage within the second handhold (panel 1b). As we have shown, unpaired regions that provide handholds can be separated by folded structures (panel 2). We also envisage that one or more of the handholds can itself be part of a folded structure (panel 3). There also appears to be flexibility in the selection of unpaired regions for handholds. We have described in this study examples where the annealing of a complementary oligonucleotide (black bar) to an unpaired region in which cleavage would otherwise have occurred does not block cleavage per se, but results in it occurring within an adjacent unpaired site (panel 4). We have also described examples where it appears that cleavage within a particular unpaired region can be facilitated by any one of several additional handholds. Thus, blocking (as illustrated) or removing a handhold does not necessarily prevent cleavage (panel 5). Should cleavage occur first within the most 3′ of two unpaired regions used for binding, it is envisaged that cleavage of the most 5′ region will be facilitated in a subsequent step by RNase E using another unpaired region as a handhold (panel 6) or perhaps a 5′-monophosphorylated end as a foothold (not shown). The unpaired region that serves as a handhold (panel 6) although shown to be located upstream of the region that is cleaved could be located downstream. The actual sequences of unpaired regions used in binding are likely to determine, at least in part, which is cleaved preferentially (65,66).
