Figure 2.
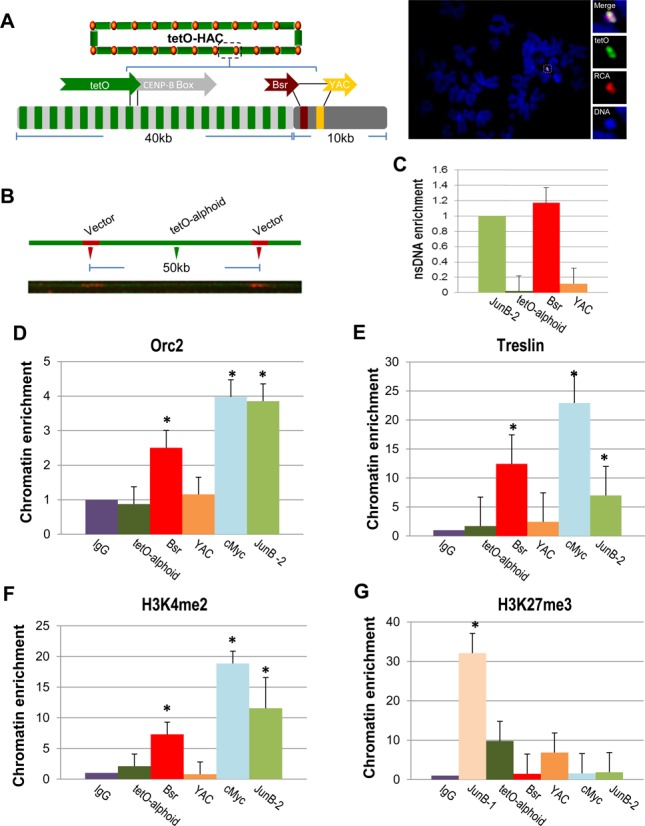
Mapping of replication initiation events within the alphoidtetO-HAC sequence. (A) Schematic of the HAC organization. The circular alphoidtetO-HAC was assembled from input DNA comprising ∼10 kb of RCA vector sequence (BAC/YAC) containing the Bsr gene and ∼40 kb of the synthetic tetO-alphoid array. The synthetic array is composed by tetO alphoid monomer (in green) arrange within every other monomer from chromosome 17 comprising CENP-B box (in gray). FISH demonstrates the presence of an autonomous form of alphoidtetO-HAC in HT1080 cells. The HAC was visualized using a vector probe (RCA) (in red) and tetO-alphoid probe (in green). (B) DNA fiber-FISH analysis using a YAC/BAC vector probe demonstrating a regular structure of the HAC, i.e. a 10 kb YAC/BAC vector signals (in red) are flanked with a 40 kb alphoidtetO array (in green). (C) Real-time PCR-based nascent DNA enrichment assay to map origins of replication along the HAC sequence. (D, E) ChIP-qPCR with antibodies against Orc2 and Treslin shows enrichment of ORC on the Bsr sequences. JUNB-2 and cMyc were used as positive controls. (F) Chromatin profiling of the alphoidtetO-HAC shows H3K4me2 accumulation on the Bsr sequences. JUNB-2 and cMyc were used as positive controls. (G) ChIP analysis of H3K27me3 chromatin in the HAC. Student's t-test was used: P < 0.01 (*), ±SD.
