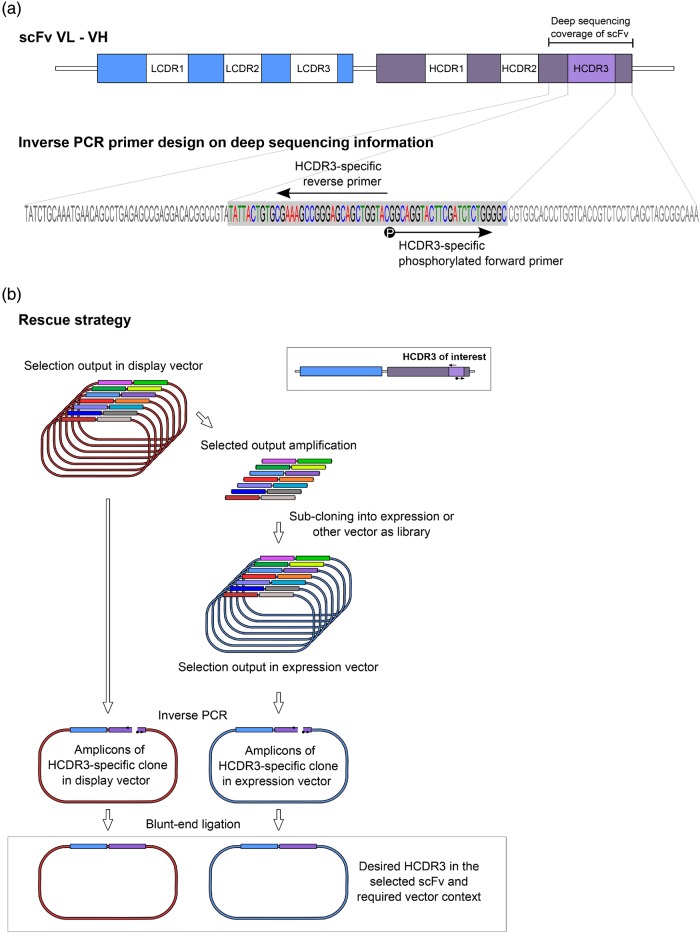
Fig. 1.
Inverse PCR strategy applied to antibodies. (a) The structure of the scFv gene. The variable light (VL) and heavy (VH) chain CDRs are indicated by white boxes. The portion of the VH covered by deep sequencing is identified and magnified: the HCDR3, as identified by the AbMining tool, is boxed in gray and the inverse PCR primers are shown. A black circle on the forward primer indicates phosphorylation. (b) Schematic representation of the specific HCDR3 rescue strategy: the desired HCDR3-specific back-to-back primers are designed on the HCDR3 sequence as obtained from deep sequencing. Plasmid DNA obtained from the selection output is used as a template for inverse PCR allowing the isolation of the desired scFv molecule identified by in vitro selection. The amplification is carried out on the selected output either in its original display vector context (left) or after subcloning into a suitable expression vector (right). The final product is a plasmid carrying the specific scFv.