Figure 1. Gor effector identification and interactome mapping.
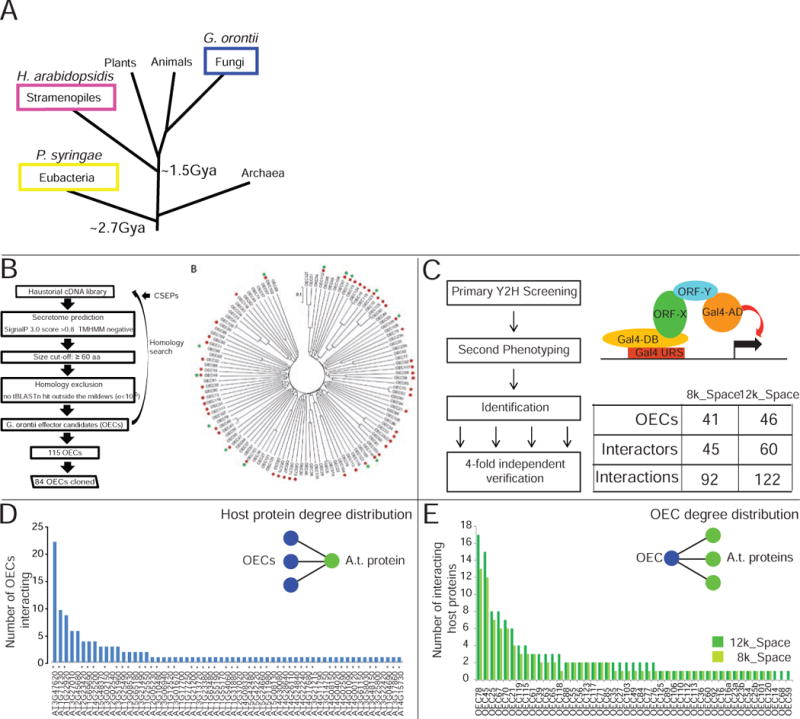
A. Golovinomyces orontii is a pathogenic ascomycete that diverged approximately 2.7 and 1.5 billion years ago (Gya), respectively, from the other Arabidopsis pathogens (Kemen and Jones, 2012; Markow, 2005). B. Effector identification pipeline and family relationships of identified and cloned Gor effector candidates (OEC) and the presence of homologs in the powdery mildews Blumeria graminis f. sp. hordei (green dots) and Erysiphe pisi (red dots). C. Our Y2H pipeline consists of three interrogation steps: screening, phenotyping and four-fold verification, resulting in the indicated number of interactions. D. Degree distribution of Arabidopsis proteins interacting with OECs. Asterisks indicate 8k_space proteins. E. Degree distribution of OECs interacting with Arabidopsis proteins in the 8k_space (light green) and 12K_space (dark green). See also Figure S1, Table S1.
