Abstract
Purpose:
To determine mechanisms by which SCCRO5 (aka DCUN1D5) promotes oncogenesis.
Experimental Design:
SCCRO5 mRNA and protein expression were assessed in 203 randomly selected primary cancer tissue samples, matched histologically normal tissues, and cell lines by use of real-time PCR and Western blot analysis. SCCRO5 overexpression was correlated with survival. The effect of SCCRO5 knockdown on viability was assessed in selected cancer cell lines. Structure-function studies were performed to determine the SCCRO5 residues required for binding to the neddylation components, for neddylation-promoting activity, and for transformation.
Results:
In oral and lung squamous cell carcinomas, SCCRO5 mRNA levels corresponded with protein levels and overexpression correlated with decreased disease-specific survival. Knockdown of SCCRO5 by RNAi resulted in a selective decrease in the viability of cancer cells with high endogenous levels, suggesting the presence of oncogene addiction. SCCRO5 promoted cullin neddylation while maintaining conserved reaction processivity paradigms involved in ubiquitin and ubiquitin-like protein conjugation, establishing it as a component of the neddylation E3. Neddylation activities in vitro required the potentiating of neddylation (PONY) domain but not the nuclear localization sequence (NLS) domain. In contrast, both the NLS domain and the PONY domain were required for transformation of NIH-3T3 cells.
Conclusions:
Our data suggest that SCCRO5 has oncogenic potential and that it imparts its oncogenicity as a component of the neddylation E3. Neddylation activity and nuclear localization of SCCRO5 are important for its in vivo function.
Keywords: DCUN1D5, oncogene, neddylation, ubiquitination, squamous cell carcinoma
Introduction
Several important anticancer therapies have been discovered by leveraging the knowledge that changes in protein homeostasis drive human cancer pathogenesis. In this regard, the ubiquitin/proteasome system is of significance, as there is a high prevalence of aberrations among the components of this pathway (1, 2). In particular, the activity of cullin-RING type ubiquitination E3 ligases is commonly dysregulated in human cancers (3). Dysregulation often results from aberrations in the substrate recognition adaptors (ie, FBX7, SPOP, Skp2, VHL), and rarely from abnormalities in core components (eg, Cul4A amplification in breast cancer) (4-7). It has long been established that posttranslational modification of cullins by neddylation is a key regulator of cullin-RING ligase (CRL) activity (neddylation promotes assembly of the CRL complex and enhances recruitment of ubiquitin-charged E2, to facilitate ubiquitin conjugation) (8-13). As is the case with ubiquitination, neddylation results from the sequential enzymatic activity of a dedicated E1 (APPBP1-Uba3), E2 (Ubc12 [aka Ube2M] and Ube2F), and E3 (14). The link between abnormal CRL activity and dysregulation of the proteins involved in neddylation in human cancers has only recently been appreciated (15). These observations have led to the development of a small molecule inhibitor of neddylation E1, MLN4924, which has shown promise in preclinical and early human trials (3).
SCCRO (aka DCUN1D1), a key component of the E3 for neddylation, is activated by amplification in a wide range of human cancers (16, 17). The role that SCCRO plays in cancer pathogenesis has been studied in both in vivo and in vitro experimental systems, establishing its function as an oncogene (16, 18). In addition, overexpression of SCCRO in human tumor samples has been independently associated with poor survival outcomes (16). Of interest, SCCRO is a member of a protein family that contains four other highly conserved paralogues in higher organisms. All family members have a conserved C-terminal potentiating of neddylation (PONY) domain with a variable N-terminal region. SCCRO paralogues are subdivided into three subfamilies on the basis of the N-terminal sequence: SCCRO and SCCRO2 (aka DCUN1D2) contain a ubiquitin-associated (UBA) domain, SCCRO3 (aka DCUN1D3) contains a myristoylation sequence, and SCCRO4 (aka DCUN1D4) and SCCRO5 (aka DCUN1D5) contain a nuclear localization sequence (NLS). We have previously shown that SCCRO is oncogenic and that its oncogenic function requires its neddylation activity (16, 17). SCCRO3 has also been shown to play a role in oncogenesis (19). Recent work indicates that SCCRO5 has oncogenic activity (20). However, the precise mechanisms by which the NLS-containing SCCRO paralogues promote oncogenesis remain unknown.
Like SCCRO (at 3q26), its paralogues are located in chromosomal loci that are recurrently amplified in human cancers (SCCRO2 at 13q34, SCCRO4 at 4q12, and SCCRO5 at 11q22) (21-23). Of these, 11q22 amplification is the second most frequent, occurring in a wide range of human cancers, with the highest prevalence in squamous cell carcinomas (SCCs) of the cervix, esophagus, head and neck, and lung (24-27). Moreover, there is an independent association between 11q22 amplification and worse clinical outcome, suggesting that this locus harbors genes that play a role in cancer pathogenesis (15, 28, 29). Although several candidate genes have been identified, including MMPs, Birc2 (cIAP1), Birc3 (cIAP2), and Yap, none has been clearly established as a target that drives selection for 11q22 amplification (29-31). Fine-mapping of 11q22.2-q22.3 in cervical, oral, and lung cancers has shown that SCCRO5 (MGC2714) is located within the minimal common region of amplification (27, 30, 32). SCCRO5 is upregulated in laryngeal SCC and putatively plays a role in its pathogenesis (20). Given the established role of SCCRO in oncogenesis, we questioned whether SCCRO5 drives selection for 11q22 amplification in human cancers and sought to elucidate the mechanisms involved.
Here, we report the validation of SCCRO5 as a target that drives selection for 11q22 amplification in human cancers. We found that SCCRO5 mRNA expression is associated with a corresponding increase in protein levels and is correlated with decreased disease-specific survival in oral and lung SCCs. Specific knockdown of SCCRO5 in cancer cell lines with high endogenous levels of SCCRO5 expression resulted in a significantly higher decrease in viability, compared with that in cells with low expression levels, suggesting the presence of an oncogene addiction phenotype. The oncogenic potential of SCCRO5 is underscored by its ability to transform fibroblasts (NIH-3T3 cells) in vitro. Furthermore, we show that, like SCCRO, SCCRO5 functions as a component of the E3, promoting cullin neddylation while maintaining reaction processivity paradigms. Although only the PONY domain was required for SCCRO5’s neddylation function in vitro, its oncogenic activity in vivo requires both the PONY domain and the NLS domain, suggesting that subcellular localization plays a role in its function. The precise mechanism involved in SCCRO5’s in vivo neddylation activity remains to be defined.
Materials and Methods
Bioinformatic analyses
The ClustalW program (Conway Institute UCD, Dublin, Ireland) was used for multiple sequence alignments. Data from The Cancer Genome Atlas (TCGA) projects were accessed and analyzed using the cBio Cancer Genomics Portal (http://www.cbioportal.org).
Human tissues, cell lines, and antibodies
A total of 203 randomly selected primary cancer tissue and adjacent matched histologically normal tissue samples were obtained from patients undergoing surgical treatment at Memorial Sloan-Kettering Cancer Center between January 1, 2004, and October 1, 2007. All patients undergoing surgical resection of primary cancers were offered an opportunity to participate in the study. Only previously untreated patients with adequate tissue stored in our tissue bank were included in the study. Informed consent was obtained from all patients, in accordance with the guidelines established by the institutional review board, and use of patient samples was approved by the institutional Human Biospecimen Utilization Committee. Histopathologic confirmation of all specimens was established by an experienced pathologist. Tissues were frozen in liquid nitrogen immediately after resection and stored at −80°C until use. Demographic and tumor data for the oral SCC, lung SCC, thyroid cancer, and lung neuroendocrine carcinoma patients are described in Supplementary Tables S1-S4.
The origins, cytogenetic characteristics, maintenance, and growth of the cell lines used in this study were as previously described (16, 33). The following antibodies were used in this study: anti-SCCRO (developed and validated as previously described [16]), anti-SCCRO5 (developed and validated in our laboratory [data not shown]), anti-Cul1 (Invitrogen, Grand Island, NY), anti-Cul2 (Abcam, Cambridge, MA), anti-Cul3 (BD Biosciences, San Jose, CA), anti-ROC1 (Abcam), anti-Ubc12 (Rockland, Gilbertsville, PA), anti-CAND1 (BD Biosciences, San Jose, CA), anti-α-tubulin and anti-GAPDH (Millipore, Billerica, MA), and anti-actin and secondary antibodies conjugated to horseradish peroxidase (Santa Cruz Biotechnology, Santa Cruz, CA).
RNA isolation, cDNA synthesis, and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from cell lines and tissue by use of Trizol reagent (Life Technologies, Grand Island, NY), repurified using RNeasy Mini spin columns (Qiagen, Valencia, CA), and treated with DNase I to eliminate residual genomic DNA. One microgram of total RNA was reverse-transcribed using MultiScribe Reverse Transcriptase (Applied Biosystems, Foster City, CA). Primers were designed for SCCRO5 and GAPDH (Supplementary Table S5), by use of the Primer3 program (Howard Hughes Medical Institute, Chevy Chase, MD), to generate PCR products 75–300 bp in length and were obtained from a commercial source (Operon Technologies, Alameda, CA). To confirm specificity for SCCRO5, the SCCRO5 primer set was tested on plasmids for all SCCRO paralogues. qRT-PCR for SCCRO5 was performed, at least in duplicate, in two separate experiments, on the 203 tumor and paired normal samples and on appropriate controls by use of a Sequence Detector System 7900HT device (Applied Biosystems, Foster City, CA), as previously described (16). To ensure the presence of a single PCR product, melting curve analysis was performed after each experiment. A standard curve was generated using serial dilutions of cDNA from MDA686 and MDA1186 cell lines. The comparative threshold cycle method was used to calculate the SCCRO5 expression ratio in each sample relative to the value observed in the control standard curve, with GAPDH used as a control for normalization among samples (34). GAPDH was determined to be the most stable housekeeping gene in the tissues studied, on the basis of geNorm analysis (M < 0.298) (35, 36). Overexpression was defined as at least 2-fold higher levels of mRNA expression in the tumor sample relative to the matched normal sample and the mean of all normal tissue samples for each cancer type.
Mutation profiling
Primers were designed, using Primer3, to cover putative exon regions of SCCRO5 (National Center for Biotechnology Information Human Genome Build 36.1), yielding products ≤500 bp in length (Supplementary Table S6) (37). Tumor DNA was extracted using the DNeasy kit (Qiagen, Valencia, CA) and was subjected to whole-genome amplification, as previously described (38). High-throughput bidirectional dideoxynucleotide sequencing of PCR-amplified gene products was performed using the Sanger sequencing platform, as previously described (39). PolyPhred and PolyScan software were used to generate an initial automated report of sequence variations (39). For each respective exon, tumor sequences were compared against reference sequences listed in the National Center for Biotechnology Information (RefSeq) database. After visual inspection of the individual forward and reverse chromograms, for confirmation of nonsynonymous sequence variations and insertions or deletions (including duplications), a manual review list of potential nucleotide changes was produced. Synonymous variants and those with corresponding entries in the Single Nucleotide Polymorphism Database (http://www.ncbi.nlm.nih.gov/projects/SNP/) were excluded from analysis.
cDNA cloning, mutagenesis, and short hairpin RNA (shRNA) knockdown
Human SCCRO5 cDNA was obtained from Clontech Laboratories (Clontech, Mountain View, CA). Mutations were generated in SCCRO5 by standard PCR mutagenesis, as previously described (17). Primers were designed using Primer3 (Supplementary Table S7). SCCRO5 and its mutated forms were cloned into pEGFP-N2 (Clontech, Mountain View, CA), pBABE (BD Biosciences, San Jose, CA), and pGEX-4T-3 (GE Healthcare Life Sciences, Piscataway, NJ) vectors. Plasmid and protein purifications were performed as previously described (16, 17). shRNA against human SCCRO5 (obtained from the Genome Sequencing Center at Memorial Sloan-Kettering Cancer Center) or lacZ shRNA (negative control) was transfected—along with packaging plasmids pLP1, pLP2, and pVSVG—into HEK293 cells by use of Lipofectamine 2000, in accordance with the manufacturer’s protocol (Invitrogen, Grand Island, NY). Lentiviral particles were harvested 48 h and 72 h after transfection and were filtered, pelleted, and resuspended in media for retroviral delivery in the presence of polybrene (Millipore, Billerica, MA). Twenty-four hours after infection, cells were cultured in media containing puromycin, to select for infected cells.
Molecular and biochemical assays
Glutathione S-transferase (GST) pull-down assays, in vitro neddylation assays, in vitro thioester reactions, transfections, and immunoblotting were performed as previously described (16, 17).
Proliferation and colony formation assays
MTS proliferation assays and colony formation assays were performed as previously described (16, 17). For the soft agar assay, colonies were counted using ImageJ software (National Institutes of Health, Bethesda, MD), to analyze images captured from four 6-well plate wells per construct, from two independent experiments.
Statistical analysis
Descriptive statistics were used to summarize study day. Continuous variables were compared across groups using Student’s t-test or Mann-Whitney U-test. Categorical comparisons were performed using the Fisher’s exact test. Survival curves were generated using the Kaplan-Meier method and compared using the log-rank test. Time to recurrence was defined as the time between completion of primary treatment of cancer and the development of recurrent cancer at any site (local, regional, or distant). Survival outcomes were censored for patients who died of causes other then cancer, were lost to follow-up or survived to the end of the study. Statistical significance was defined as a two tailed p-value less than or equal to 0.05. All statistical analyses were performed using state-of-the-art statistical packages (SPSS, Chicago, IL; or SAS, Durham, NC).
Results
SCCRO5 binds to neddylation components
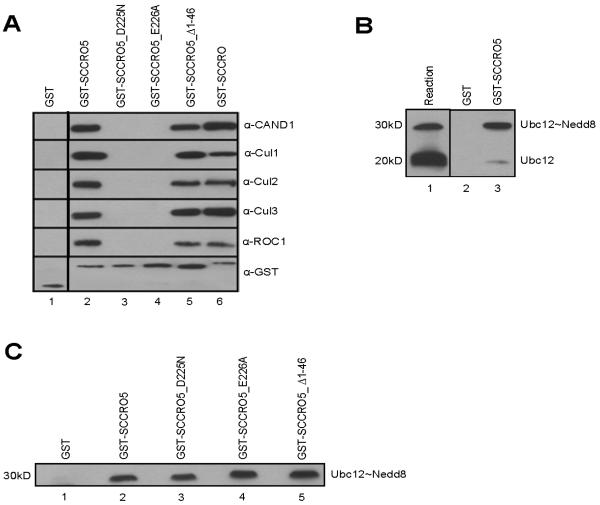
In silico analysis showed that SCCRO has four paralogues in higher organisms, all with a highly conserved C-terminal PONY domain with variable N-terminal domains (Supplementary Fig. S1A). The established role of the PONY domain in neddylation raised the possibility that, like SCCRO, SCCRO’s paralogues participate in neddylation E3 activity. To determine the role that SCCRO5 plays in neddylation, we assessed the binding of SCCRO5 to neddylation components. GST pull-down assays using HeLa cell extracts, followed by Western blot analysis for neddylation components, showed that, like SCCRO, SCCRO5 binds to CAND1, Ubc12, Cul1, Cul2, Cul3, and ROC1 (Table 1 and Fig. 1A). Previous biochemical and structural studies have established that SCCRO binds to its neddylation components through its PONY domain, which contains four residues that are critical for binding interactions (Supplementary Fig. S1B) (17, 40, 41). To map the binding domains, we created a series of SCCRO5 deletions and point mutants as GST fusions, including those involving the N-terminal NLS and C-terminal PONY domains, and used them in pull-down assays on HeLa lysates. Deletion of the PONY domain (SCCRO5_Δ189-237) or mutation in critical residues (SCCRO5_D195N, SCCRO5_A219R, SCCRO5_D225N, SCCRO5_E226A, SCCRO5_A219R/D225N, and SCCRO5_D195N/A219R/D225N) resulted in loss of binding to CAND1 and Cul1-ROC1 (Table 1). Binding to Ubc12 was lost only with larger deletion of the C-terminal region of SCCRO5 (SCCRO5_Δ189-237), which is consistent with the location of the binding site of Ubc12 in SCCRO (17). Previous studies have shown that the UBA domain in SCCRO is not required for binding to neddylation components (17, 40, 41). SCCRO5 contains an NLS in its N-terminus. We found that the NLS is functional, as transiently expressed GFP-SCCRO5 was exclusively present in the nucleus, whereas NLS-deletion mutant GFP-SCCRO5 (SCCRO5_Δ1-10 and SCCRO5_Δ1-46) had a pan-cellular distribution on fluorescence microscopy (data not shown). As with the UBA domain of SCCRO, deletion of the NLS or the entire N-terminus in SCCRO5 (SCCRO5_Δ1-10 and SCCRO5_Δ1-46, respectively) had no effect on binding to Cullin-ROC1, Ubc12, or CAND1 (Table 1). These results show that binding with the neddylation components and residues involved in the interactions are conserved between SCCRO and SCCRO5.
TABLE 1.
SCCRO5 interacts with components of the neddylation pathway.
| SCCRO5 Mutants | Cul1-ROC1 | CAND1 | Ubc12 |
|---|---|---|---|
| WT | + | + | + |
| Δ1-10 | + | + | + |
| Δ1-46 | + | + | + |
| Δ189-237 | − | − | − |
| A219R | − | − | + |
| D195N | − | − | + |
| D225N | − | − | + |
| E226A | − | − | − |
| A219R/ D225N | − | − | + |
| D195N/ A219R/ D225N | − | − | + |
Table showing a summary of results from pull-down assays using GST-SCCRO5 and selected mutants to recombinant Cul1-ROC1, Ubc12, and CAND1, which confirmed that binding to Cul1-ROC1 and CAND1 requires an intact PONY domain but not the NLS domain. In contrast, binding to Ubc12 was only lost with deletion of the C-terminus (SCCRO5_Δ189-237).
FIGURE 1.
SCCRO5 interacts with components of the neddylation pathway. A, Western blot analysis of the products of GST-SCCRO, GST-SCCRO5, and selected GST-SCCRO5 mutants after pull-down assays from HeLa lysates probed with the indicated antibodies, which shows that, like SCCRO, SCCRO5 binds to CAND1, Cul1, Cul2, Cul3, and ROC1. Mutations in the PONY domain (SCCRO5_D225N and SCCRO5_E226A), but not the N-terminal NLS (SCCRO5_Δ1-46), result in loss of binding. The dividing line between lane 1 and lane 2 indicates the position of omitted lanes from the same gel. B, Western blot analysis of Ubc12 after GST and GST-SCCRO5 pull-down assays on products from a thioester reaction (lane 1), which shows preferential binding to Ubc12~Nedd8, even in the presence of excess free Ubc12. C, Western blot analysis of Ubc12 after pull-down assays on products of thioester reaction, which shows that GST-SCCRO5 and PONY and NLS mutants bind equally to Ubc12~Nedd8.
SCCRO5 preferentially binds to Ubc12~Nedd8 thioester
The mechanisms that maintain the processivity of the reactions that result in the conjugation of ubiquitin and ubiquitin-like proteins are highly conserved. Central to maintenance of processivity is a differential affinity between E1 and E3 for free and conjugated E2, respectively. Differences in affinity ensures that an E2 must dissociate from its cognate E1 after it accepts transfer of ubiquitin or ubiquitin-like proteins and before it can bind to the E3. SCCRO has greater affinity for E2~Nedd8 thioester than for free E2, which is consistent with its role as a component of the E3 for neddylation (17). To determine whether SCCRO5 also conforms to the established processivity paradigm for E3s, we performed GST pull-down assays on products from a Ubc12 thioester reaction, followed by Western blot analysis for Ubc12. Even in the presence of molar-excess free Ubc12, SCCRO5 preferentially bound to Ubc12~Nedd8 thioester (Fig. 1B [compare lanes 1 and 3]). Moreover, the results of GST pull-down assays showed that, as with SCCRO, critical residues involved in binding of SCCRO5 to Cul1-ROC1 (lanes 3 and 4) and the N-terminal region (lane 5) were not involved in to Ubc12~Nedd8 (Fig. 1C). These findings confirm that, in its interactions with Ubc12, SCCRO5 maintains reaction processivity paradigms.
SCCRO5 promotes cullin neddylation
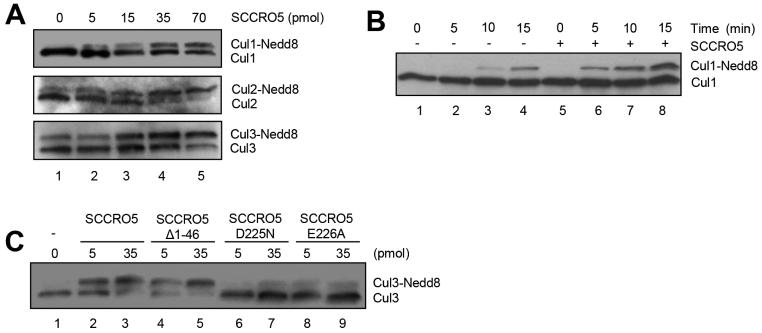
Structural and biochemical studies have shown that SCCRO promotes cullin neddylation by promoting complex assembly, nuclear translocation of neddylation E3 components, enhanced E2 recruitment, and structural orientation to optimize the efficiency of the transfer of Nedd8 from Ubc12 to cullin (42, 43). To determine whether SCCRO5 also promotes cullin neddylation, we performed in vitro neddylation reactions containing Nedd8, recombinant APPBP1/Uba3 (E1), Ubc12 (E2), ATP, and whole-cell lysate from HeLa cells (as a source of Cullin-ROC substrates), with and without SCCRO5. Western blotting of reaction products for cullins showed a dose-dependent increase in neddylation of Cul1, Cul2, and Cul3 with the addition of SCCRO5 (Fig. 2A). A time-course in vitro neddylation reaction showed that SCCRO5 enhanced the rate of cullin neddylation (Fig. 2B). To determine the domains required for the observed effects on neddylation, we supplemented in vitro reactions with SCCRO5 or selected SCCRO5 mutants. PONY domain mutants (SCCRO5_D225N and SCCRO5_E226A) failed to augment Cul3 neddylation beyond basal levels, whereas N-terminus deletion mutants (SCCRO5_Δ1-46) enhanced Cul3 neddylation to levels similar to those observed in reactions supplemented with wild-type SCCRO5 (Fig. 2C). Combined, these findings suggest that, as with SCCRO (17), the effect of SCCRO5 on cullin neddylation in vitro requires its interaction with neddylation E3 components through its PONY domain but not its NLS domain.
FIGURE 2.
SCCRO5 augments cullin neddylation. A, Western blot analysis of the indicated cullins on products of an in vitro neddylation reaction using HeLa lysates as a source for Cullin-ROC1 complexes supplemented with a gradient of SCCRO5, which shows a dose-dependent increase in neddylation of Cul1, Cul2, and Cul3 with the addition of recombinant SCCRO5. B, Western blot analysis of Cul1 on products of an in vitro neddylation reaction with (lanes 5-8) or without (lanes 1-4) the addition of SCCRO5, which shows enhanced efficiency of Cul1 neddylation by SCCRO5. C, Western blot analysis of Cul3 on products of an in vitro neddylation reaction with concentration gradients of SCCRO5, SCCRO5_Δ1-46, SCCRO5_D225N, or SCCRO5_E226A (quantities in pmol), which shows a dose-dependent increase in neddylation of Cul3 with SCCRO5 and SCCRO5_Δ1-46 but not with SCCRO5_D225N or SCCRO5_E226A.
SCCRO5 promotes cell proliferation and anchorage-independent cell growth
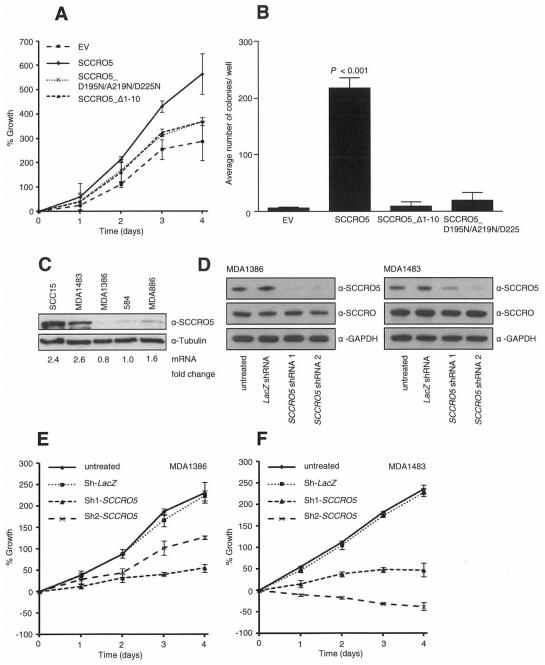
With SCCRO5’s function in cullin neddylation established, we next asked whether this activity is required for transformation. To determine this, we transfected NIH-3T3 cells with pBABE-SCCRO5, selected pBABE-SCCRO5 mutants or empty vector, and developed two stably expressing clones for each construct by use of puromycin selection. Equal expression of SCCRO5 and SCCRO5 mutants in selected clones was confirmed by qRT-PCR and Western blot analysis (Supplementary Fig. S2A and S2B). The results of MTS assays showed that the proliferation rate was significantly higher in SCCRO5-expressing clones than in SCCRO5_D195N/A219R/D225N, SCCRO5_Δ1-10, or empty vector transfected NIH-3T3 cells (Fig. 3A). NIH-3T3 clones were then subjected to soft agar colony formation assay to assess transforming ability, which showed that SCCRO5-transfected clones had significantly higher anchorage-independent growth, compared with SCCRO5_D195N/A219R/D225N, SCCRO5_Δ1-10, or empty vector transfected cells (Fig. 3B). Combined, these findings suggest that SCCRO5’s oncogenic activity requires its neddylation-promoting activity, as well as its compartmentalization to the nucleus.
FIGURE 3.
Transgenic expression of SCCRO5 promotes proliferation and anchorage-independent growth. A, Graph showing results from an MTS assay on NIH-3T3 cells stably expressing SCCRO5 and indicated mutants, which shows increased proliferation in plasmids expressing SCCRO5 but not NLS (SCCRO5_Δ1-10) or PONY (SCCRO5_D195N/A219N/D225N) domain mutants. B, Results from a soft agar assay, which shows increased colony formation in NIH-3T3 cells stably transfected with SCCRO5, compared with that in cells transfected with empty vector and SCCRO5 NLS and PONY domain mutants (bars represent the mean ± SD number of colonies per well of 6-well plates; P < 0.001). C, Western blot analysis, which shows SCCRO5 protein levels and corresponding mRNA levels, on the basis of real-time PCR, in head and neck cancer cell lines. D, Western blot analysis of SCCRO5, with GAPDH as a loading control, on lysates from MDA1386 (low endogenous SCCRO5) and MDA1483 (high endogenous SCCRO5), before and after transfection of two independent shRNA constructs against SCCRO5 or lacZ control, which shows efficient and specific knockdown of SCCRO5. E and F, Graphs from MTS assays showing a more pronounced decrease in the viability of MDA1483 cells, compared with MDA1386 cells, with knockdown of SCCRO5.
SCCRO5 overexpression is associated with an oncogene addiction phenotype
Consistent with the concept of oncogene addiction, we previously reported that cancer cell lines with amplification and high levels of SCCRO expression are more susceptible to apoptosis with RNAi knockdown of SCCRO, compared with cell lines with normal copy numbers and low levels of SCCRO expression (16, 44). To validate the role that SCCRO5 plays in cancer pathogenesis, we sought to determine whether an oncogene addiction to SCCRO5 was present in cancer cell lines. Results of qRT-PCR and Western blot analysis showed a wide range of SCCRO5 expression in a panel of head and neck cancer cell lines, with the highest levels of expression in SCC15 and MDA1483 and the lowest level in MDA1386 (Fig. 3C). Representative cell lines with low (MDA1386) and high (MDA1483) endogenous SCCRO5 expression were transfected with two independent anti-SCCRO5 shRNA constructs and with anti-lacZ shRNA as a control. Western blot analysis, after transfection with anti-SCCRO5 shRNA constructs, showed efficient knockdown of SCCRO5 levels, compared with those in the anti-lacZ shRNA transfected cells (Fig. 3D). SCCRO levels were not affected by transfection with any of the shRNA constructs, which confirmed specificity. Results of MTS assays showed a more significant decrease in the viability of MDA1483 cells, compared with MDA1386 cells, after SCCRO5 knockdown (Fig. 3E and 3F). These results indicate the presence of an oncogene addiction phenotype and validate the position that SCCRO5 plays a role in cancer pathogenesis.
SCCRO5 overexpression is associated with an aggressive clinical course
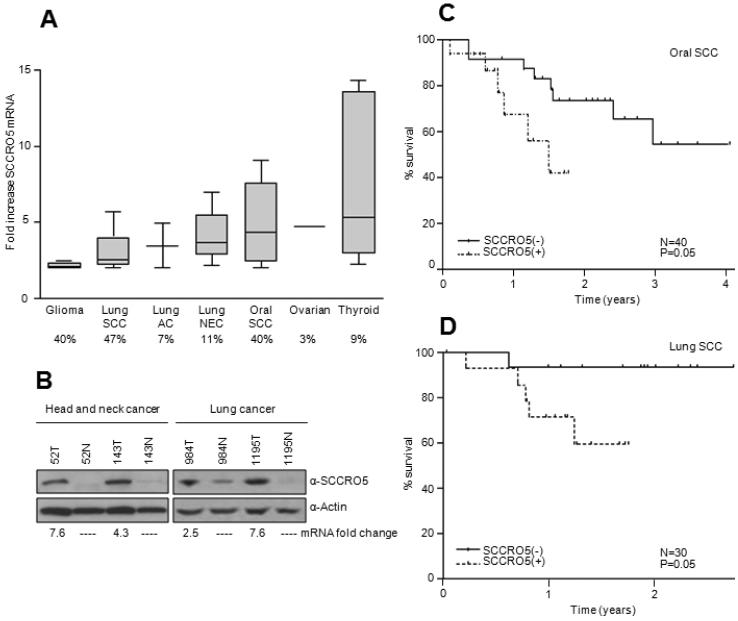
To establish the clinical significance of SCCRO5 overexpression, we sought to identify mutations and to determine the prevalence of SCCRO5 overexpression, as well as the association of mutations and overexpression with outcome in a panel of randomly selected human tumors. Sequenome-based mutational screening of all cases did not identify any mutations in SCCRO5 (data not shown). To investigate the presence and frequency of overexpression, SCCRO5 mRNA levels were determined by qRT-PCR in cancer tissue and matched histologically normal tissue for each case. The highest prevalence of SCCRO5 mRNA overexpression was observed in cancer types known to harbor 11q22 amplification, including gliomas (4 of 10 [40%]), lung SCCs (14 of 30 [47%]), and oral cavity SCCs (16 of 40 [40%]). SCCRO5 mRNA overexpression was less common in cancer types with a lower prevalence of 11q22 amplification, including lung adenocarcinomas (2 of 27 [7%]), lung neuroendocrine carcinomas (6 of 54 [11%]), ovarian carcinomas (1 of 40 [3%]), and thyroid carcinomas (5 of 56 [9%]) (Fig. 4A) (26, 27, 30). Moreover, in the cohort of lung SCCs and oral cavity SCCs, SCCRO5 mRNA levels correlated with SCCRO5 protein levels (Fig. 4B).
FIGURE 4.
SCCRO5 overexpression is common in human tumors and correlates with outcomes. A, Box plot showing fold increase in SCCRO5 mRNA expression, analyzed by qRT-PCR, in 10 gliomas, 30 lung squamous cell carcinomas (SCCs), 27 lung adenocarcinomas (ACs), 54 lung neuroendocrine (NEC) tumors, 40 oral SCCs, 40 ovarian carcinomas, and 56 thyroid carcinomas, compared with matched normal tissues (boxes represent the lower through the upper quartile; the median is shown as a horizontal line; whiskers represent minimum and maximum levels). The percentage of cases with overexpression is given below each plot. B, Western blot analysis showing SCCRO5 protein expression in representative head and neck and lung SCCs (T) and matched normal (N) samples. The corresponding fold change in mRNA levels, determined by real-time PCR, is noted below. C, Kaplan-Meier survival curves from post-hoc analysis showing recurrence-free survival based on SCCRO5 mRNA expression status in primary oral SCCs. D, Kaplan-Meier survival curves showing recurrence-free survival from post-hoc analysis based on SCCRO5 mRNA expression status in primary lung SCCs.
To validate the prevalence of SCCRO5 dysregulation, we analyzed interim results from TCGA projects, which showed that the highest prevalence of SCCRO5 dysregulation (amplification and/or overexpression) was in head and neck SCCs (11.6%), followed by ovarian serous cystadenocarcinomas (3%; copy number change only; expression data not available), cervical carcinomas (8.7%; copy number change only; expression data not available), and bladder urothelial carcinomas (7%), with a significant correlation between copy number and mRNA expression in head and neck SCCs (Supplementary Fig. S3). The difference in the prevalence of dysregulation between our tumor cohort and that in the TCGA likely represents differences in the sensitivity of the analytic techniques used to assess SCCRO5 mRNA levels (cDNA array vs. qRT-PCR) and in the type of control tissue used for analysis (matched normal tissue vs. blood).
To determine clinical relevance, we performed a post hoc analysis to determine the association between SCCRO5 overexpression and disease-free survival. We limited survival comparisons to patients undergoing treatment for head and neck or lung SCC as these data sets had : (1) reasonable power (more then 10 cases with SCCRO5 over expression) (2) had sufficient events (only 2 recurrences were observed in the thyroid cancer cohorts; Supplementary Table 3); (3) had uniform histology (thyroid cohort included papillary, follicular and Hürthle cell carcinomas and lung neuroendocrine carcinomas included large cell carcinomas, small cell carcinomas, and carcinoid tumors; Supplementary Table 4). All patients in the study cohort underwent uniform treatment that included primary surgery with or without adjuvant treatment (on the basis of the extent of disease), in accordance with established institutional protocols. Clinicopathologic parameters, including age, sex, tumor node metastasis stage, and tobacco use, were not significantly different according to SCCRO5 expression status for either cancer cohort (Supplementary Tables S1 and S2). We found that SCCRO5 overexpression (SCCRO5+), which is defined in the Methods section, compared with normal expression of SCCRO5 (SCCRO5−), negatively correlated with disease-free survival for both head and neck SCCs and lung SCCs (Fig. 4C and 4D; P = 0.05). Interestingly, time to survival analysis for lung neuroendocrine carcinomas also shows worse outcome in tumors with SCCRO5 over expression (Supplementary Figure 4; P = 0.05). However, the differences in outcome in the lung neuroendocrine carcinoma need to be considered with caution given the limited sample size and variable behavior of histological subtypes of neuroendocrine carcinomas. Moreover, limitation of sample size and number of events in the study cohorts do not support multivariate analyses. Nonetheless, the correlation of overexpression with an aggressive clinical course supports SCCRO5’s clinical relevance.
Discussion
SCCRO and its paralogues are commonly dysregulated in human cancers, with aberrations of one or more paralogues present in 50% of head and neck cancers, in 70% of lung SCCs, in 55% of ovarian serous cystadenocarcinomas, in 34% of cervical and endometrial carcinomas, in 26% of lung adenocarcinomas, and in 15% of glioblastomas, on the basis of analysis of interim data from respective TCGA projects. Of the SCCRO paralogues, SCCRO and SCCRO5 are the most commonly dysregulated and have a tendency toward mutual exclusivity, which suggests that they act independently in oncogenesis. It is well established that overexpression of SCCRO promotes its function as an oncogene (16, 18). By showing that overexpression of SCCRO5 has transforming activity and by associating it with an aggressive clinical course in human cancers, our data support the position that SCCRO5 may function as an oncogene as well (20). Its role in cancer pathogenesis is further validated by our findings, which indicate the presence of an oncogene addiction phenotype in cancer cell lines that have high endogenous levels of SCCRO5 expression.
Structure-function analyses show that, as with SCCRO, SCCRO5’s conserved PONY domain is required for binding to neddylation components, as well as for its neddylation and transforming activities (16, 17, 41, 42). Of interest, SCCRO5’s N-terminal NLS was not required for neddylation-promoting activity in vitro, which is similar to the case for the UBA domain in SCCRO (41). Given that SCCRO’s subcellular localization is important to its neddylation activity, the direct or indirect association of the N-terminal motifs with subcellular localization of the SCCRO paralogues suggests that they may regulate neddylation activity in vivo (43-45). Consistent with this finding, SCCRO’s UBA is involved in monoubiquitination by Nedd4-1, which promotes nuclear export, thereby inhibiting its neddylation activity (43, 45) (unpublished data). Similarly, the myristoylation sequence localizes SCCRO3 to the membrane, which affects its neddylation promoting activity (46) (unpublished data). Our finding showing that the NLS is required for SCCRO5’s transforming activity suggests that, as with the UBA, the NLS may modulate neddylation activity in vivo. However, these findings raise a question about SCCRO5’s role in neddylation. Our previous work showed that SCCRO promotes nuclear translocation of Cullin-ROC1, which is required for neddylation in vivo. Given that SCCRO5 is present almost exclusively in the nucleus, it is unclear how cytoplasmic Cullin-ROC1 complexes are delivered to the nucleus to allow for interaction with SCCRO5. Several possibilities exist, including cooperative activity between other SCCRO paralogues and NLS-containing variants, presence of alternative mechanisms for nuclear translocation of Cullin-ROC1 complexes, and selective activity on cullins situated in the nucleus.
The functional effects of neddylation are mediated by downstream targets that CRLs activate. The recent publication by Monda and colleagues showing that SCCRO and its paralogues have overlapping affinity to E2s (Ube2M and Ube2F) and cullins (42) suggests that they may have redundant effects on CRL activity in vivo. This idea is supported by the differential requirements of SCCRO for viability in yeast, whose genome does not contain other SCCRO paralogues, and in higher organisms, where paralogues can compensate for SCCRO loss (40, 43) (unpublished data). Since SCCRO5 promotes neddylation of all cullins (with the possible exception of Cul4A), it putatively can regulate the ubiquitination of a myriad of proteins, resulting in diverse cellular effects (42). Guo and colleagues suggest that SCCRO5 may be involved in regulating ubiquitination of the proteins involved in DNA-damage repair (20). Consistent with this suggestion, SCCRO5 was found to be part of a panel of seven genes whose expression score predicts radiation response in patients with cervical cancer (47). Further work is required to define the CRLs and protein targets that are dependent on SCCRO5 in the DNA-damage response, cellular activities, and oncogenesis.
Supplementary Material
Translational relevance.
The importance of the ubiquitin-proteasome pathway in cancer pathogenesis is established by the therapeutic efficacy of agents that target pathway activity in treating human cancers. Neddylation, a major regulator of ubiquitin-proteasome pathway activity, is itself subject to regulation by SCCRO. SCCRO is commonly activated by amplification in many different types of cancer and its overexpression is independently associated with poor clinical outcome. Here we show that SCCRO5, a paralogue of SCCRO, plays a role in cancer pathogenesis. SCCRO5 is commonly overexpressed in human cancers and its overexpression is associated with worse outcome in oral and lung squamous cell carcinomas. Establishing its role in cancer pathogenesis, we show oncogene addiction associated with SCCRO5 overexpression in cancer cell lines. Moreover, we show that SCCRO5’s oncogenic activity requires it function as an E3 in neddylation, as well as, its subcellular localization. Combined our findings suggest that like SCCRO, SCCRO5 is a putative therapeutic target in oral and lung SCC, as well as, other types of cancer.
Acknowledgments
We thank Dr. Nicholas Socci, for help with analysis of gene expression data; Dr. Adriana Heguy, for help with mutation analysis; and David Sewell for excellent editorial assistance.
Financial support: This work was supported, in part, by grants from Catherine van Tussenbroek Fonds and the foundation Vrijvrouwe van Renswoude (to C.C.B.); Fulbright, The Dutch Cancer Society, and the Prins Bernhard Culture Foundation (to V.B.W.); and the Clinical Innovator Award from the Flight Attendants Medical Research Institute (to B.S.).
Footnotes
Conflicts of interest: None disclosed
References
- 1.Liu Z, Zanata SM, Kim J, Peterson MA, Di Vizio D, Chirieac LR, et al. The ubiquitin-specific protease USP2a prevents endocytosis-mediated EGFR degradation. Oncogene. 2013;32:1660–9. doi: 10.1038/onc.2012.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grim JE, Knoblaugh SE, Guthrie KA, Hagar A, Swanger J, Hespelt J, et al. Fbw7 and p53 cooperatively suppress advanced and chromosomally unstable intestinal cancer. Mol Cell Biol. 2012;32:2160–7. doi: 10.1128/MCB.00305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soucy TA, Dick LR, Smith PG, Milhollen MA, Brownell JE. The NEDD8 conjugation pathway and its relevance in cancer biology and therapy. Genes Cancer. 2010;1:708–16. doi: 10.1177/1947601910382898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsu JM, Lee YC, Yu CT, Huang CY. Fbx7 functions in the SCF complex regulating Cdk1-cyclin B-phosphorylated hepatoma up-regulated protein (HURP) proteolysis by a proline-rich region. J Biol Chem. 2004;279:32592–602. doi: 10.1074/jbc.M404950200. [DOI] [PubMed] [Google Scholar]
- 5.Lehman NL, van de Rijn M, Jackson PK. Screening of tissue microarrays for ubiquitin proteasome system components in tumors. Methods Enzymol. 2005;399:334–55. doi: 10.1016/S0076-6879(05)99023-X. [DOI] [PubMed] [Google Scholar]
- 6.Chen LC, Manjeshwar S, Lu Y, Moore D, Ljung BM, Kuo WL, et al. The human homologue for the Caenorhabditis elegans cul-4 gene is amplified and overexpressed in primary breast cancers. Cancer Res. 1998;58:3677–83. [PubMed] [Google Scholar]
- 7.Melchor L, Saucedo-Cuevas LP, Munoz-Repeto I, Rodriguez-Pinilla SM, Honrado E, Campoverde A, et al. Comprehensive characterization of the DNA amplification at 13q34 in human breast cancer reveals TFDP1 and CUL4A as likely candidate target genes. Breast Cancer Res. 2009;11:R86. doi: 10.1186/bcr2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen A, Wu K, Fuchs SY, Tan P, Gomez C, Pan ZQ. The conserved RING-H2 finger of ROC1 is required for ubiquitin ligation. J Biol Chem. 2000;275:15432–9. doi: 10.1074/jbc.M907300199. [DOI] [PubMed] [Google Scholar]
- 9.Kawakami T, Chiba T, Suzuki T, Iwai K, Yamanaka K, Minato N, et al. NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 2001;20:4003–12. doi: 10.1093/emboj/20.15.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng J, Yang X, Harrell JM, Ryzhikov S, Shim EH, Lykke-Andersen K, et al. CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol Cell. 2002;10:1519–26. doi: 10.1016/s1097-2765(02)00784-0. [DOI] [PubMed] [Google Scholar]
- 11.Goldenberg SJ, Cascio TC, Shumway SD, Garbutt KC, Liu J, Xiong Y, et al. Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell. 2004;119:517–28. doi: 10.1016/j.cell.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 12.Sakata E, Yamaguchi Y, Miyauchi Y, Iwai K, Chiba T, Saeki Y, et al. Direct interactions between NEDD8 and ubiquitin E2 conjugating enzymes upregulate cullin-based E3 ligase activity. Nat Struct Mol Biol. 2007;14:167–8. doi: 10.1038/nsmb1191. [DOI] [PubMed] [Google Scholar]
- 13.Wimuttisuk W, Singer JD. The Cullin3 ubiquitin ligase functions as a Nedd8-bound heterodimer. Mol Biol Cell. 2007;18:899–909. doi: 10.1091/mbc.E06-06-0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong L, Yeh ET. Identification of the activating and conjugating enzymes of the NEDD8 conjugation pathway. J Biol Chem. 1999;274:12036–42. doi: 10.1074/jbc.274.17.12036. [DOI] [PubMed] [Google Scholar]
- 15.Zhao Y, Sun Y. Cullin-RING ligases (CRLs) as attractive anti-cancer targets. Curr Pharm Des. 2012 doi: 10.2174/13816128113199990300. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarkaria I, P O-c, Talbot SG, Reddy PG, Ngai I, Maghami E, et al. Squamous cell carcinoma related oncogene/DCUN1D1 is highly conserved and activated by amplification in squamous cell carcinomas. Cancer Res. 2006;66:9437–44. doi: 10.1158/0008-5472.CAN-06-2074. [DOI] [PubMed] [Google Scholar]
- 17.Kim AY, Bommelje CC, Lee BE, Yonekawa Y, Choi L, Morris LG, et al. SCCRO (DCUN1D1) is an essential component of the E3 complex for neddylation. J Biol Chem. 2008;283:33211–20. doi: 10.1074/jbc.M804440200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Broderick SR, Golas BJ, Pham D, Towe CW, Talbot SG, Kaufman A, et al. SCCRO promotes glioma formation and malignant progression in mice. Neoplasia. 2010;12:476–84. doi: 10.1593/neo.10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma T, Shi T, Huang J, Wu L, Hu F, He P, et al. DCUN1D3, a novel UVC-responsive gene that is involved in cell cycle progression and cell growth. Cancer Sci. 2008;99:2128–35. doi: 10.1111/j.1349-7006.2008.00929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo W, Li GJ, Xu HB, Xie JS, Shi TP, Zhang SZ, et al. In vitro biological characterization of DCUN1D5 in DNA damage response. Asian Pac J Cancer Prev. 2012;13:4157–62. doi: 10.7314/apjcp.2012.13.8.4157. [DOI] [PubMed] [Google Scholar]
- 21.Abba MC, Fabris VT, Hu Y, Kittrell FS, Cai WW, Donehower LA, et al. Identification of novel amplification gene targets in mouse and human breast cancer at a syntenic cluster mapping to mouse ch8A1 and human ch13q34. Cancer Res. 2007;67:4104–12. doi: 10.1158/0008-5472.CAN-06-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salgado R, Toll A, Alameda F, Baro T, Martin-Ezquerra G, Sanmartin O, et al. CKS1B amplification is a frequent event in cutaneous squamous cell carcinoma with aggressive clinical behaviour. Genes Chromosomes Cancer. 2010;49:1054–61. doi: 10.1002/gcc.20814. [DOI] [PubMed] [Google Scholar]
- 23.Barrow J, Adamowicz-Brice M, Cartmill M, MacArthur D, Lowe J, Robson K, et al. Homozygous loss of ADAM3A revealed by genome-wide analysis of pediatric high-grade glioma and diffuse intrinsic pontine gliomas. Neuro Oncol. 2011;13:212–22. doi: 10.1093/neuonc/noq158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lockwood WW, Chari R, Chi B, Lam WL. Recent advances in array comparative genomic hybridization technologies and their applications in human genetics. Eur J Hum Genet. 2006;14:139–48. doi: 10.1038/sj.ejhg.5201531. [DOI] [PubMed] [Google Scholar]
- 25.Brown J, Bothma H, Veale R, Willem P. Genomic imbalances in esophageal carcinoma cell lines involve Wnt pathway genes. World J Gastroenterol. 2011;17:2909–23. doi: 10.3748/wjg.v17.i24.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ambatipudi S, Gerstung M, Gowda R, Pai P, Borges AM, Schaffer AA, et al. Genomic profiling of advanced-stage oral cancers reveals chromosome 11q alterations as markers of poor clinical outcome. PLoS One. 2011;6:e17250. doi: 10.1371/journal.pone.0017250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dai Z, Zhu WG, Morrison CD, Brena RM, Smiraglia DJ, Raval A, et al. A comprehensive search for DNA amplification in lung cancer identifies inhibitors of apoptosis cIAP1 and cIAP2 as candidate oncogenes. Hum Mol Genet. 2003;12:791–801. doi: 10.1093/hmg/ddg083. [DOI] [PubMed] [Google Scholar]
- 28.Muramatsu T, Imoto I, Matsui T, Kozaki K, Haruki S, Sudol M, et al. YAP is a candidate oncogene for esophageal squamous cell carcinoma. Carcinogenesis. 2011;32:389–98. doi: 10.1093/carcin/bgq254. [DOI] [PubMed] [Google Scholar]
- 29.Wu C, Xu B, Yuan P, Miao X, Liu Y, Guan Y, et al. Genome-wide interrogation identifies YAP1 variants associated with survival of small-cell lung cancer patients. Cancer Res. 2010;70:9721–9. doi: 10.1158/0008-5472.CAN-10-1493. [DOI] [PubMed] [Google Scholar]
- 30.Baldwin C, Garnis C, Zhang L, Rosin MP, Lam WL. Multiple microalterations detected at high frequency in oral cancer. Cancer Res. 2005;65:7561–7. doi: 10.1158/0008-5472.CAN-05-1513. [DOI] [PubMed] [Google Scholar]
- 31.Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–67. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lockwood WW, Coe BP, Williams AC, MacAulay C, Lam WL. Whole genome tiling path array CGH analysis of segmental copy number alterations in cervical cancer cell lines. Int J Cancer. 2007;120:436–43. doi: 10.1002/ijc.22335. [DOI] [PubMed] [Google Scholar]
- 33.Singh B, Gogineni S, Goberdhan A, Sacks P, Shaha A, Shah J, et al. Spectral karyotyping analysis of head and neck squamous cell carcinoma. Laryngoscope. 2001;111:1545–50. doi: 10.1097/00005537-200109000-00010. [DOI] [PubMed] [Google Scholar]
- 34.Essentials of real time PCR [Internet] Applied Biosystems. 2012 Available from: http://www3.appliedbiosystems.com/cms/groups/mcb_marketing/documents/generaldocuments/cms_039996.pdf.
- 35.Gene Quantification [Internet] Reference genes/housekeeping genes. Available from: http://www.gene-quantification.de/hkg.html.
- 36.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–86. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 38.Little SE, Vuononvirta R, Reis-Filho JS, Natrajan R, Iravani M, Fenwick K, et al. Array CGH using whole genome amplification of fresh-frozen and formalin-fixed, paraffin-embedded tumor DNA. Genomics. 2006;87:298–306. doi: 10.1016/j.ygeno.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 39.Marks JL, McLellan MD, Zakowski MF, Lash AE, Kasai Y, Broderick S, et al. Mutational analysis of EGFR and related signaling pathway genes in lung adenocarcinomas identifies a novel somatic kinase domain mutation in FGFR4. PLoS One. 2007;2:e426. doi: 10.1371/journal.pone.0000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurz T, Chou YC, Willems AR, Meyer-Schaller N, Hecht ML, Tyers M, et al. Dcn1 functions as a scaffold-type E3 ligase for cullin neddylation. Mol Cell. 2008;29:23–35. doi: 10.1016/j.molcel.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 41.Kurz T, Ozlu N, Rudolf F, O'Rourke SM, Luke B, Hofmann K, et al. The conserved protein DCN-1/Dcn1p is required for cullin neddylation in C. elegans and S. cerevisiae. Nature. 2005;435:1257–61. doi: 10.1038/nature03662. [DOI] [PubMed] [Google Scholar]
- 42.Monda JK, Scott DC, Miller DJ, Lydeard J, King D, Harper JW, et al. Structural conservation of distinctive N-terminal acetylation-dependent interactions across a family of mammalian NEDD8 ligation enzymes. Structure. 2013;21:42–53. doi: 10.1016/j.str.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang G, Kaufman AJ, Ramanathan Y, Singh B. SCCRO (DCUN1D1) promotes nuclear translocation and assembly of the neddylation E3 complex. J Biol Chem. 2011;286:10297–304. doi: 10.1074/jbc.M110.203729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weinstein IB, Joe A. Oncogene addiction. Cancer Res. 2008;68:3077–80. doi: 10.1158/0008-5472.CAN-07-3293. discussion 3080. [DOI] [PubMed] [Google Scholar]
- 45.Wu K, Yan H, Fang L, Wang X, Pfleger C, Jiang X, et al. Mono-ubiquitination drives nuclear export of the human DCN1-like protein hDCNL1. J Biol Chem. 2011;286:34060–70. doi: 10.1074/jbc.M111.273045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meyer-Schaller N, Chou YC, Sumara I, Martin DD, Kurz T, Katheder N, et al. The human Dcn1-like protein DCNL3 promotes Cul3 neddylation at membranes. Proc Natl Acad Sci U S A. 2009;106:12365–70. doi: 10.1073/pnas.0812528106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rajkumar T, Vijayalakshmi N, Sabitha K, Shirley S, Selvaluxmy G, Bose MV, et al. A 7 gene expression score predicts for radiation response in cancer cervix. BMC Cancer. 2009;9:365. doi: 10.1186/1471-2407-9-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.