Fig. 3.
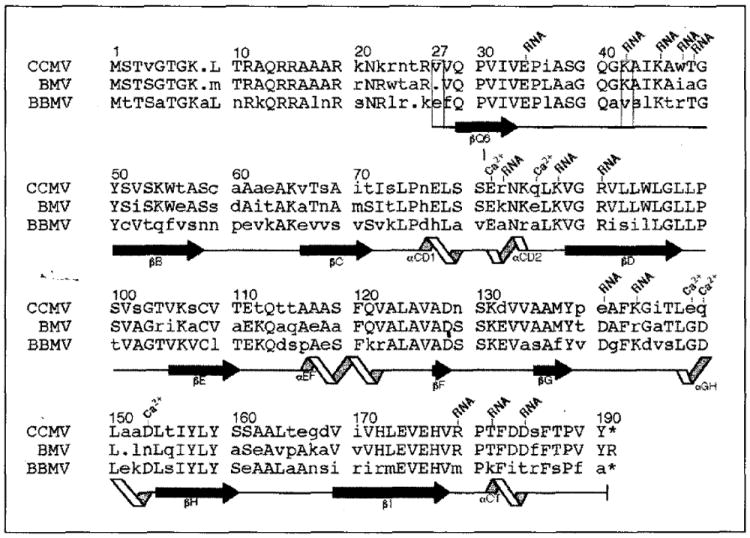
Sequence alignment of the bromovirus group capsid proteins and designation of the corresponding polypeptide secondary structure (CCMV, cowpea chlorotic mottle virus; BMV, bromegrass mosaic virus; BBMV, broad bean mosaic virus) [4,57,58]. Residue numbering at the top of each sequence block is for the CCMV protein with the left-most digit over the column to which the number corresponds. All other labels are based upon analysis of the high resolution CCMV capsid structure. Upper-case letters represent residues which are identical in at least two of the three sequences. The capsid proteins of CCMV and BMV share 70% identity whereas CCMV and BBMV are only 48% identical. Depending on the local capsid environment, ordered electron density for residues 27 (first box) to 190 (B and C subunits) or for residues 42 (second box) to 190 (A subunits) is observed. Residues which are involved in RNA binding and calcium ion coordination (Ca2+) are labeled. Secondary-structure elements determined by the program PROCHECK [59] are represented at the bottom of each sequence block. Extended black arrows and twisting ribbons represent β-sheet and α-helix secondary structure, respectively. These symbols are placed under the residues to which the represented structure has been assigned, and each labeled according to their location in the CCMV protein tertiary fold (Fig. 2a). Solid lines between β-sheets and α-helices have no regular secondary structure assigned. The dots indicate inserted gaps for purposes of alignment and the asterisks indicate that the equivalent terminal residues are not present in CCMV and BBMV.
