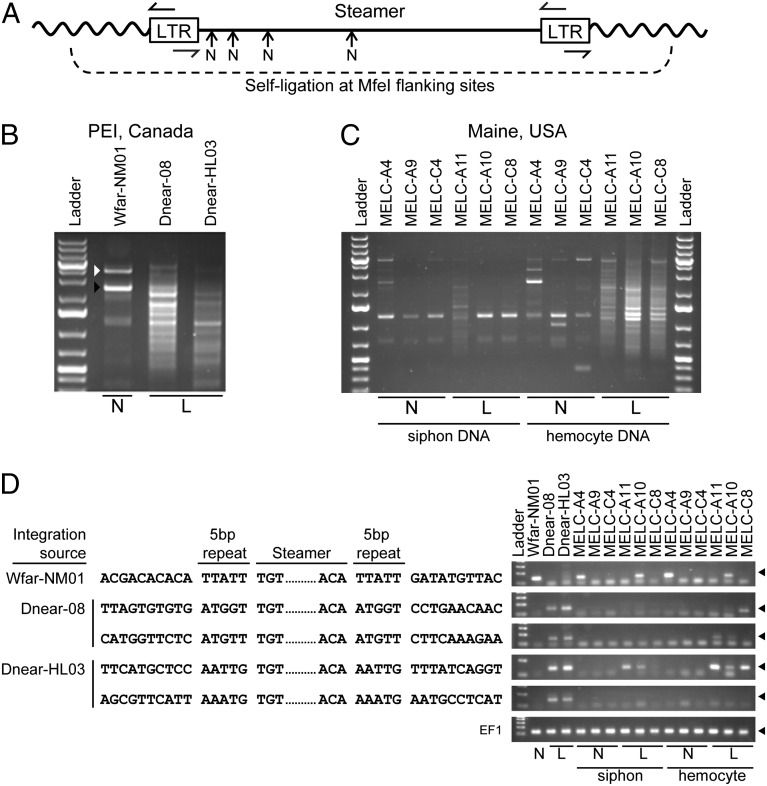
Fig. 5.
Identification of Steamer integration sites in normal and diseased clams. Inverse PCR was used to clone and sequence integration sites of the Steamer retroelement. (A) Schematic of inverse PCR methodology: genomic DNA was digested with MfeI (cleaving only in the flanking DNA), circularized by ligation, and redigested with NsiI at internal sites (N), and finally PCR was performed with outward-directed LTR primers. (B) Inverse PCR was performed using mantle tissue DNA of one normal animal (N), and hemocyte DNA of two heavily leukemic animals (L) from PEI, Canada, and the PCR products were analyzed by agarose gel electrophoresis. For WfarNM01, the white arrowhead marks amplification of the internal Steamer sequence (due to incomplete NsiI cleavage) and the black arrowhead marks the junction product of a single Steamer copy. The leukemic samples yielded a large number of heterogeneous junction products. (C) Inverse PCR was performed using DNA from both siphon tissue and hemocytes for normal and leukemic animals from Maine, USA. (D) Representative DNA sequences of individual cloned integration sites from normal (one site) and leukemic DNAs (two sites from each leukemic PEI animal) are shown. The genomic DNA flanking sequences, the 5-bp duplicated repeats, and the Steamer termini are shown. The presence of the integration sites in the source DNAs and in DNA from other animals was determined for each of the sequences shown by a diagnostic PCR using a forward primer in the Steamer LTR and a reverse primer in the flanking genomic DNA (Right; products are ∼150 bp and marked with a filled triangle). Amplification of EF1 is included as a control.