Figure 5. Effect of transcript elongation.
(A) Active elongation sequesters core RNAPs for the length of the operon and sigma subunit for some nucleotides. (B) Formation of holoenzymes in the presence of one type of sigma factor without DNA (no specific binding and no transcription with 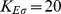 nM, dashed line), in the presence of specific binding (holoenzymes bind to promoter with
nM, dashed line), in the presence of specific binding (holoenzymes bind to promoter with 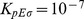 M but do not transcribe, case (i)) and in the presence of both specific binding and transcription (case (ii)). The black bars (
M but do not transcribe, case (i)) and in the presence of both specific binding and transcription (case (ii)). The black bars (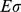 ) show the case when sigma factor and core unbind as holoenzyme (the binding affinity is described by the equilibrium dissociation constant), the dark blue (
) show the case when sigma factor and core unbind as holoenzyme (the binding affinity is described by the equilibrium dissociation constant), the dark blue ( ) and the light blue bars (
) and the light blue bars (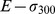 ) when sigma factor separates from core either after promoter unbinding or gene transcription and after 300 nucleotides, respectively (thus, the binding affinity is
) when sigma factor separates from core either after promoter unbinding or gene transcription and after 300 nucleotides, respectively (thus, the binding affinity is 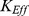 ). (C) Number of holoenzymes
). (C) Number of holoenzymes 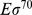 and
and  as a function of the copy number of alternative sigma factors in the absence of DNA (case (i)), with transcription of both σ
70- and σAlt-dependent genes but with unbinding of sigma factor after 300 nucleotides and core at the end of the operon (case (ii)) and only with the transcription of the σAlt-dependent genes (case (iii)). Values of the parameters are the same as in Figure 5B. (D) Formation of holoenzymes
as a function of the copy number of alternative sigma factors in the absence of DNA (case (i)), with transcription of both σ
70- and σAlt-dependent genes but with unbinding of sigma factor after 300 nucleotides and core at the end of the operon (case (ii)) and only with the transcription of the σAlt-dependent genes (case (iii)). Values of the parameters are the same as in Figure 5B. (D) Formation of holoenzymes 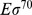 and
and 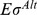 as a function of the copy number of alternative sigma factors without DNA (dashed lines) and transcript elongation (solid lines). (E) Modulation of the effective binding affinities
as a function of the copy number of alternative sigma factors without DNA (dashed lines) and transcript elongation (solid lines). (E) Modulation of the effective binding affinities 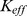 by sigma factor competition related to the case of Figure 5D. (F) Normalized transcription rate for σ
70- and σAlt-dependent promoters as a function of the number of alternative sigma factors, related to the case of Figure 5D (with
by sigma factor competition related to the case of Figure 5D. (F) Normalized transcription rate for σ
70- and σAlt-dependent promoters as a function of the number of alternative sigma factors, related to the case of Figure 5D (with 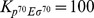 nM and
nM and 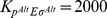 nM).
nM).

