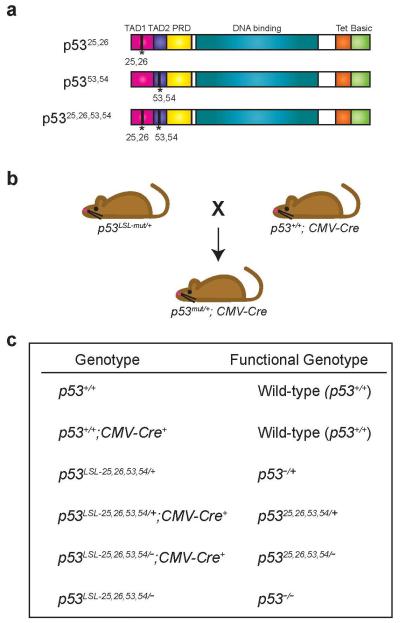
Extended Data Figure 1. Model for Examining p53-Associated Developmental Phenotypes.
(a) Schematic of p5325,26, p5353,54, and p5325,26,53,54 mutant p53 proteins. TAD: Transactivation Domain 1 or 2, PRD: Proline-Rich Domain, Tet: Tetramerization Domain, Basic: Basic Residue-Rich Domain. (b) p53LSL-mut/+ mice (where mut can denote any of the p53 TAD mutants) were crossed to p53+/+;CMV-Cre mice, which express Cre in the germline, to assess viability and developmental phenotypes of the p53 mutant-expressing progeny. (c) Table summarizing the actual genotypes and ultimate functional genotypes of progeny from crosses of p53LSL-25,26,53,54/+ and p53+/−;CMV-Cre mice, as used throughout the manuscript. While p53LSL-25,26,53,54/+;CMV-Cre is the actual initial genotype, when Cre acts to delete the Lox-Stop-Lox cassette, the genotype will be written as p5325,26,53,54/+ to reflect this recombination. In the text and figure labels, the Cre nomenclature for both control and p5325,26,53,54/+ embryos is excluded for simplicity. Controls for analyses comprise embryos both with and without the CMV-Cre transgene, as summarized in Extended-Data Fig. 3.