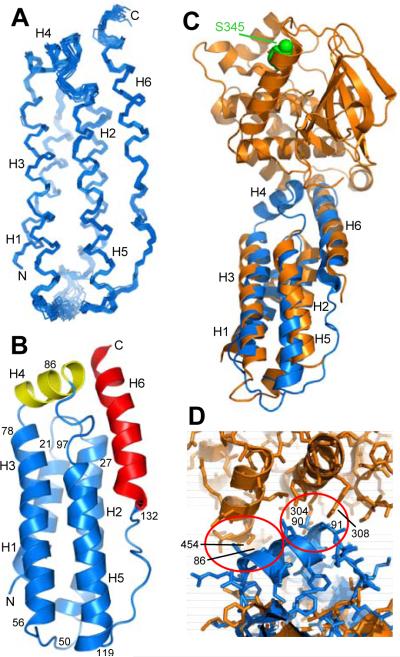
Figure 4.
Structure of MLKL(2-154) in solution. A. Backbone superposition of the 20 NMR structures of MLKL(2-154) with the lowest NOE energies. The six α-helices are labeled H1-H6 and correspond to residues 2-20 (H1), 25-47 (H2), 56-80 (H3), 82-91 (H4), 99-120 (H5) and 133-150 (H6). N and C indicate the N- and C-termini, respectively. B. Ribbon diagram of the solution structure of MLKL(2-154). The helices that form the four-helix bundle are colored in blue, the helix in the top that contains C86 (helix H4) in yellow, and the C-terminal helix (H6) in red. The positions of selected residues are indicated to help identifying the locations of residues discussed in the text within the structure. C. Superposition of ribbon diagrams of the solution structure of MLKL(2-154) (blue) and the crystal structure of mouse MLKL (orange) (PDB accession code 4BTF) (Murphy et al., 2013). The atoms of the residue of the kinase-like domain of mouse MLKL that is phosphorylated by RIP3 (S345) are shown as spheres and colored in green. D. Close-up of the interface between the kinase-like domain and the N-terminal helical domain in the structural superposition shown in panel C but showing also the proteins as stick models. The red ellipses show areas where helix H4, which is observed in our solution structure but is not observed in the crystal structure, would have steric clashes with the kinase-like domain in the relative orientation between the two domains observed in the crystal structure. Selected residues involved in the clashes are labeled.