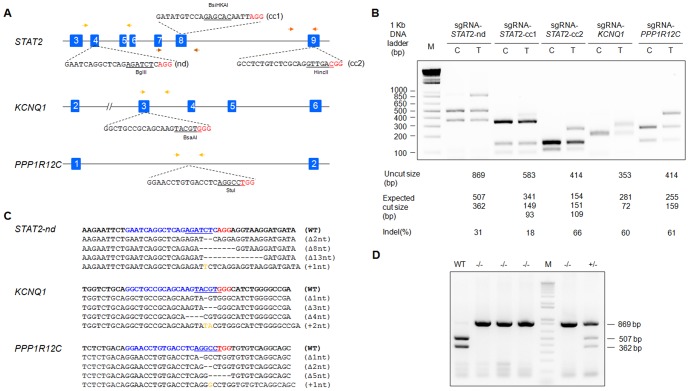
Figure 1. Gene targeting in golden Syrian hamster somatic cells by CRISPR/Cas9.
(A) Schematic diagram for the targeting sites at STAT2, KCNQ1 and PPP1R12C loci by sgRNAs. The sgRNA target sequences for each locus are depicted, with the restriction enzyme recognition sites used for the PCR-RFLP assays underlined. Letters in red are the protospacer adjacent motif (PAM) (3). Arrows: locations of PCR primers. (B) Gene targeting efficiency at the STAT2-nd, STAT2-cc1, STAT2-cc2, KCNQ1, and PPP1R12C loci in BHK cells detected by PCR-RFLP assays. M: 1 kb Plus DNA Ladder; C: controls (untransfected BHK cells); T: transfected BHK cells. (C) Indels introduced by CRISPR/Cas9 for the STAT2-nd, KCNQ1, and PPP1R12C loci in BHK cells. Letters in blue are sgRNA target sequences (with PAM in red). Nucleotide deletions and insertions are indicated by dashes and orange letters, respectively. (D) PCR-RFLP assays on single cell derived BHK cell colonies. WT: wild type; Δ1 nt, Δ2 nt, Δ3 nt, Δ4 nt, Δ5 nt, Δ8 nt, and Δ13 nt: one-, two-, three-, four-, five-, eight-, and 13-nucleotide deletion, respectively; +1 nt and +2 nt: one- and two-nucleotide insertions, respectively; -/-: both alleles targeted, and +/-: single allele targeted.