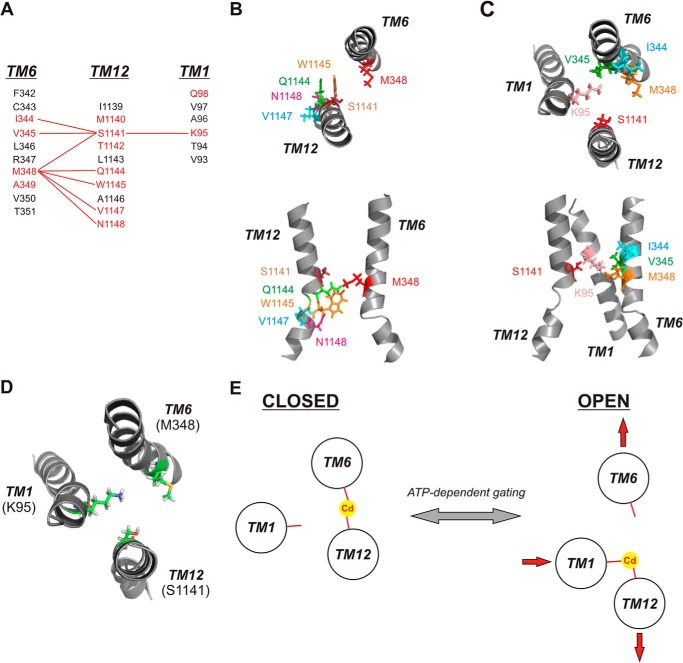
FIGURE 8.
Cadmium bridges between different TMs stabilize different channel conformations. A, identification of different Cd2+ bridges formed between different TMs. Primary sequences of TMs 1, 6, and 12 are aligned as described previously (3). (Note: On the figure, the TMs are shown in the order (left to right) 6 - 12 - 1 for visual clarity.) Residues in red are those with pore-lining side chains mutated in the present study; those in black are non-pore-lining. Red lines connect residues that can form Cd2+ bridges following mutation to cysteine. B, relative location and orientation of Met-348 (TM6) with those residues in TM12, which can form Cd2+ bridges with M348C following mutation to cysteine. C, relative location and orientation of Ser-1141 (TM12) with those residues in TMs 1 and 6, which can form Cd2+ bridges with S1141C following mutation to cysteine. In both B and C, individual TMs are illustrated as in the atomic model of Fig. 1, viewed both from the top (upper panels) and from the side (lower panels). D, relative location and orientation of key Cd2+ bridge-forming residues Lys-95 (TM1), Met-348 (TM6), and Ser-1141 (TM12), viewed from above. E, proposed graphic model for Cd2+ bridge formation between the key sites illustrated in D in open and closed states, and proposed implications for conformational rearrangement of these three TMs associated with channel gating. The closed state is stabilized by a Cd2+ bridge between M348C (TM6) and S1141C (TM12), and the open state is stabilized by a Cd2+ bridge between K95C (TM1) and S1141C (TM12). In this model, ATP-dependent channel opening and closing are associated with a relative lateral movement of different TM helices (depicted by red arrows) with no requirement for helical rotation.