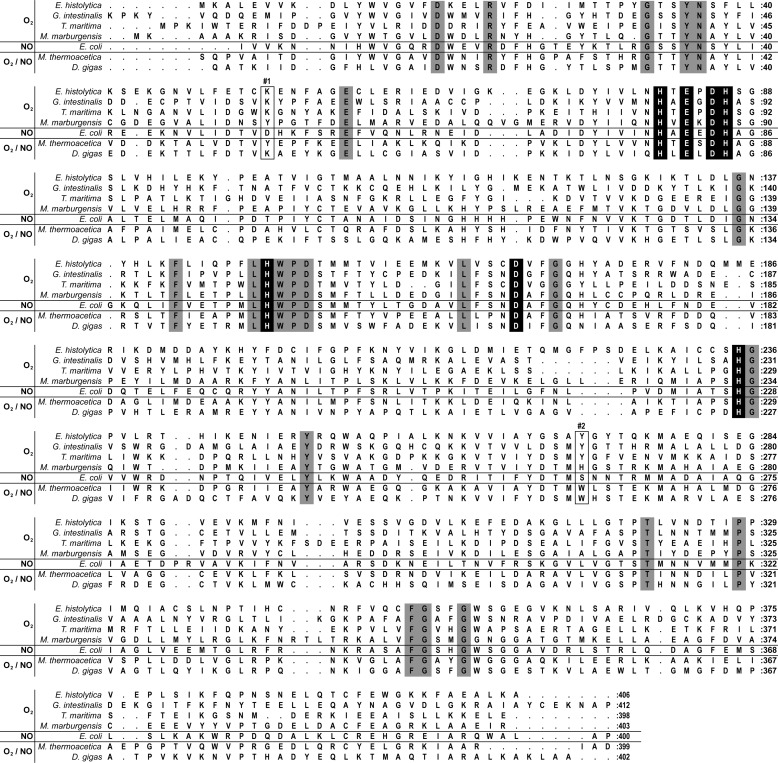
FIGURE 2.
Amino acid sequence alignment based on structural superposition of FDPs. The EhFdp1 amino acid sequence was aligned against the amino acid structural alignment built on the basis of the structures of the FDPs from G. intestinalis (PDB code 2Q9U, chain A), T. maritima (PDB code 1VME, chain A), M. marburgensis (PDB code 2OHI, chain A), E. coli FDP-D (PDB code 4D02), M. thermoacetica (PDB code 1YCF, chain C), and D. gigas (PDB code 1E5D, chain A). Gray shades indicate conserved residues; black shades correspond to diiron ligands; boxes indicate the residues in positions 1 and 2. The selectivity for O2 or NO is also indicated.