FIGURE 7.
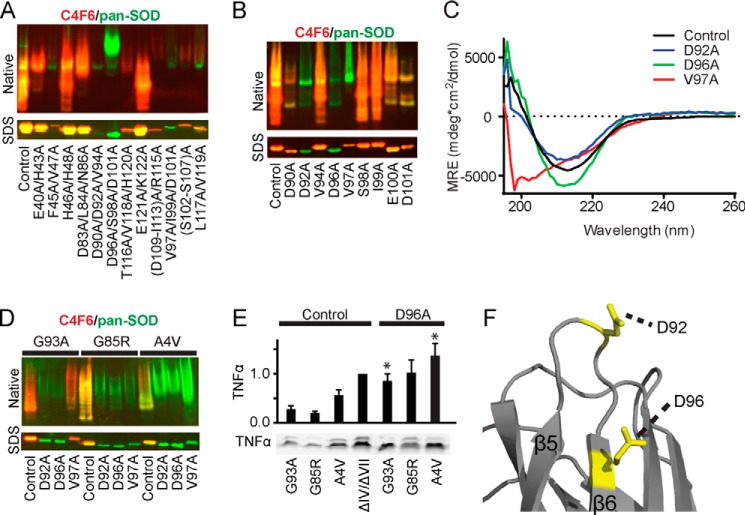
Amino acids Asp92 and Asp96 in SOD1 are required for C4F6 binding. Mutagenesis pinpoints key amino acids in misfolded SOD1 responsible for C4F6 reactivity. A, Western analyses of SOD1ΔIV/ΔVII with the indicated mutations (residue number corresponds to full-length SOD1). Pan-SOD1 (green) and C4F6 (red) antibodies were employed to assess partially purified SOD1 expression and reactivity with C4F6, respectively (control = WT SOD1ΔIV/ΔVII). Native Western analysis (top panels) shows a loss of C4F6 reactivity for the following proteins: D90A/D92A/V94A, D96A/S98A/D101A, V97A/I99A/D101A, and F45A/V47A. Denaturing Western analysis (SDS) of SOD1ΔIV/ΔVII variants reveal a loss of C4F6 reactivity for the triple mutations V97A/I99A/D101A, D90A/D92A/V94A, and D96A/S98A/D101A. B and D, the same analyses described in A demonstrate the D92A and D96A mutations induce a loss of C4F6 reactivity for SOD1ΔIV/ΔVII (B) and full-length ALS-linked SOD1 variants (D) under both native and denaturing conditions. Control indicates WT SOD1ΔIV/ΔVII in B, whereas in D, Control refers to the respective variant without the additional mutation. C, in contrast to D92A and D96A, the V97A mutation significantly perturbs the secondary structure of SOD1ΔIV/ΔVII as indicated by CD spectroscopy. MRE, mean residue ellipticity; Control, WT SOD1ΔIV/ΔVII. E, ALS-linked proteins containing the D96A mutation retain the ability to activate microglia as indicated by TNFα Western analysis (bottom panel, performed as described in Fig. 5). Quantification of TNFα is displayed above. Statistically significant comparisons between ALS-linked SOD1 variants and their D96A counterparts are indicated (*, p < 0.05; n = 2). F, the location of amino acids Asp92 and Asp96 mapped onto SOD1ΔIV/ΔVII (Protein Data Bank code 4BCZ).