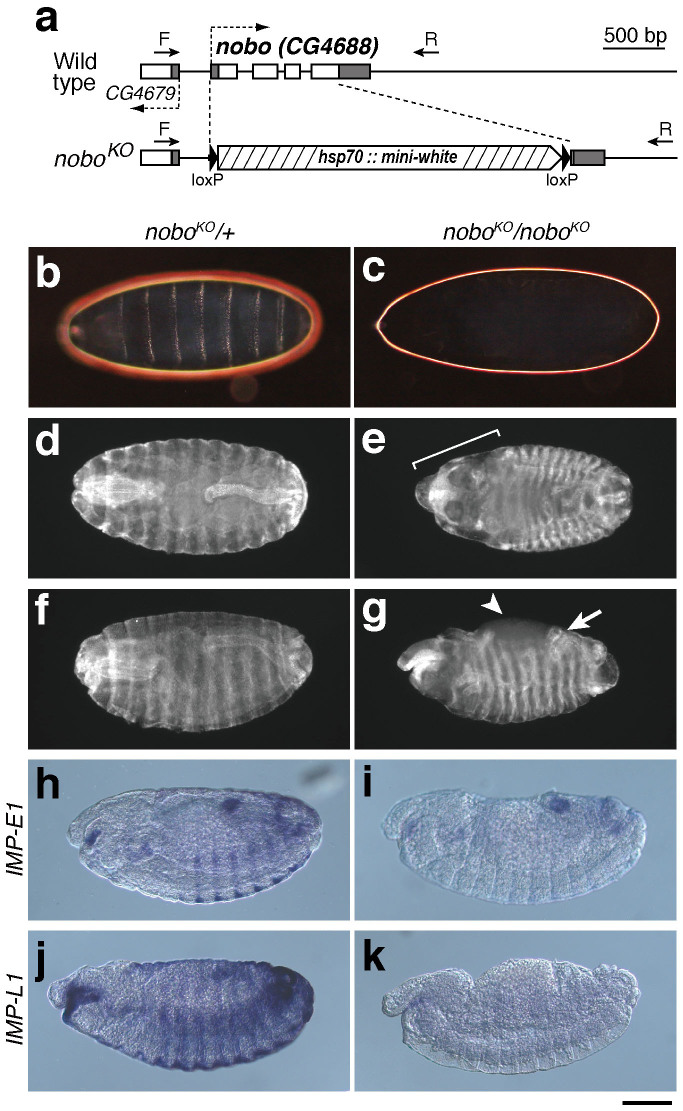
Figure 2. Generation and phenotypic analysis of noboKO homozygous mutant embryos.
(a) Genomic and exon-intron structures of the nobo (CG4688) loci of the wild-type and nobo knock-out (noboKO) strains. White and black boxes indicate the coding sequence and the untranslated regions, respectively. CG4649 is a gene located next to nobo. Dashed arrows indicate orientations of the genes. In the noboKO allele, 1,033 bp of the nobo gene region was replaced with a gene-targeting construct including the hsp70::mini-white marker cassette with loxP sites, resulting in a 680 bp deletion in the entire (696 bp) nobo coding sequence. Arrows ‘F' and ‘R' indicate the positions of the genotyping primers used in Supplementary fig. 2 (also see Methods and Supplementary table S3). (b,c) Dark-field images of embryonic cuticles from (b) noboKO heterozygous (noboKO/+) and (c) homozygous (noboKO/noboKO) embryos. (d–g) Anti-FasIII antibody staining to visualise overall embryo morphology. (d,f) noboKO/+ embryos. (e, g) noboKO/noboKO embryos. (e) The bracket indicates defective head involution. (g) The arrow and the arrowhead indicate the dorsal open phenotype and abnormal gut looping, respectively. (h–k) Expression patterns of (h, i) IMP-E1 and (j, k) IMP-L1 in stage 14 embryos. (h, j) noboKO/+ embryos. (i, k) noboKO/noboKO embryos. These data show that the nobo mutant exhibited severe reductions of these 20E-inducible genes. Scale bars: 100 µm for all images.