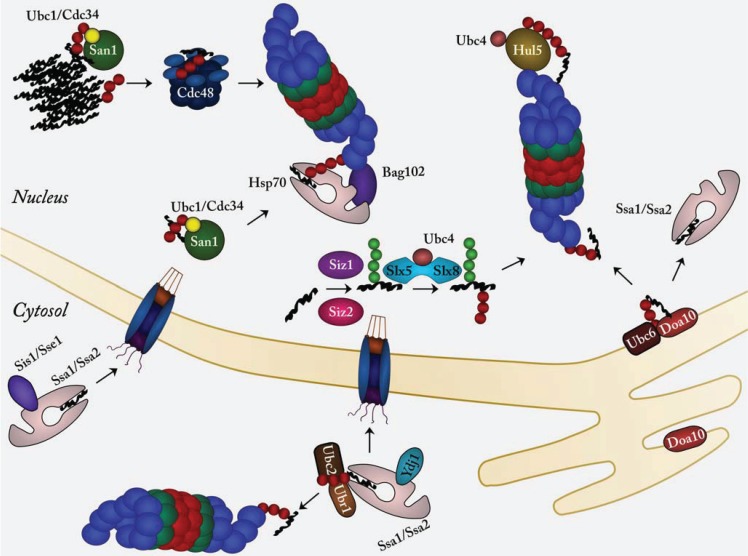
Figure 1.
Pathways for nuclear protein quality control in yeast. Conjugating ubiquitin (red sphere) to misfolded proteins (black thread) occurs in the nucleus primarily via the San1 E3 ubiquitin-protein ligase (green sphere) and its cognate E2 enzymes Ubc1 (yellow) and Cdc34 (dark red). In fission yeast the Bag102 Hsp70/proteasome co-factor links Hsp70 chaperones (pink) directly with the 26S proteasome. The transmembrane ER/nuclear envelope E3 ligase Doa10 (red) and its cognate E2 Ubc6 (brown) also target certain Ssa1 and Ssa2 Hsp70 (pink) clients for proteasomal degradation. The Slx5-Slx8 heterodimeric STUbL (blue) targets misfolded proteins that have first been marked with SUMO (green sphere) by the SUMO E3s Siz1 and Siz2. The Sis1 and Sse1 co-chaperones (purple) mediate transport of cytosolic chaperone clients to the nucleus. Due to some substrate overlap between San1 and the cytosolic Ubr1 E3 ubiquitin protein ligase, Ubr1 (light brown) and its cognate E2 Ubc2 (dark brown) may also participate in nuclear protein quality control. Finally, at the 26S proteasome the proteasome-associated E3 Hul5 (yellow), may further ubiquitylate the misfolded substrates, to ensure efficient degradation. See text for further details.