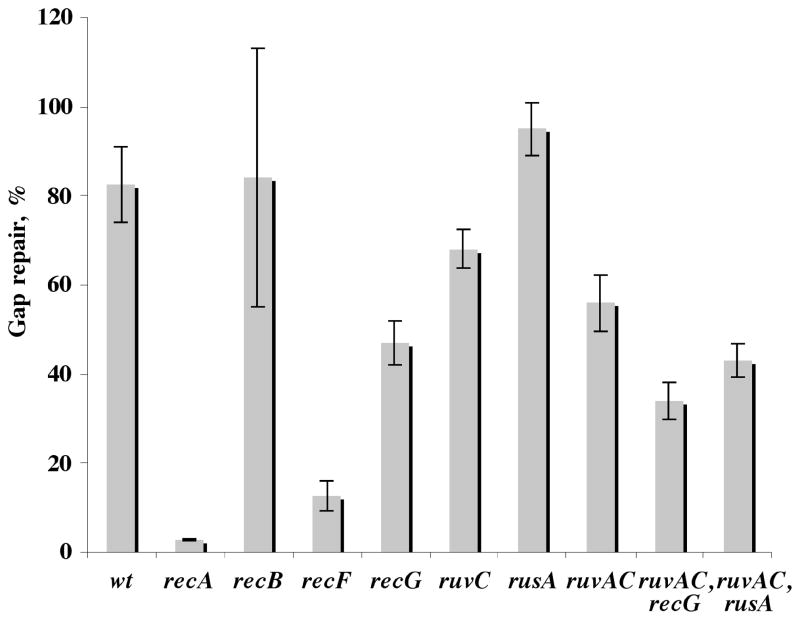
Fig. 2.
Gap-lesion plasmid repair in recombination-deficient mutants. E. coli cells were transformed with either the heterologous partner plasmid pUC18 (ampR) or the homologous partner plasmid pFGP20/Tamp (ampR). AmpR-resistant colonies were used for a second round of transformation with the gap-lesion plasmid GP21 (kanR). The same colony was used in parallel for transformation with GP20 (gap-plasmid without lesion). All strains where assayed for their survival relative to GP20 transformants. The results presented here are the average of at list three independent experiments each with at list two repetitions. The strains used are identified by genotype and are AB1157 (wt), WBN10 (ΔrecA:: tn10), WBY166 (recB21), WBY220 (ΔrecF349), WBL16 (RecG265::cat), WBL19(RuvC67:: cat) AM821 (ΔrusA::kan), AM547 (ΔruvAC65), TNM1219 (ΔruvAC65, ΔrecG263::kan), AM888 (ΔruvAC65, ΔrusA:: kan).
All strains are isogenic derivatives of E. coli AB1157 (argE3 hisG4 leuB6 proA2 ter1 ara14 galK2 lacY1 mtl1 xyl5 thi1 tsx33 rpsL31 supE44). Strains E. coli WBL16 (recG265::cat), WBL19 (ruvC67::cat), and WBL13 (mutS::Tn10) are derivatives of AB1157 constructed by generalized P1 transudation using as donors the strains N4452 (recG265::cat), N4453 (ruvC67::cat), and MM294 (mutS::Tn10), respectively. Sources of other strains were: AM547 (Δruv(A–C)65), AM821 (ΔrusA::kan), AM888 (Δruv(A–C)65, ΔrusA:: kan), and TNM1219 (Δruv(A–C)65, ΔrecG263::kan), lab collection of R. G. Lloyd; N4453 (RuvC67:: cat), E. coli genetic stock center; MM294mutS (glnV44(AS) λ-rfbC1 0 endA1 spoT1 0 thi-1 hsdR17 creC510 mutS::Tn10), WBN2 (3recA:: Tn10), WBY 100 (Δ (umuDC)595:cat), WBY166 (recB21 ArgA:Tn10), and WBY220 (ΔrecF349 TnaA:Tn10), lab collection of Z. Livneh.