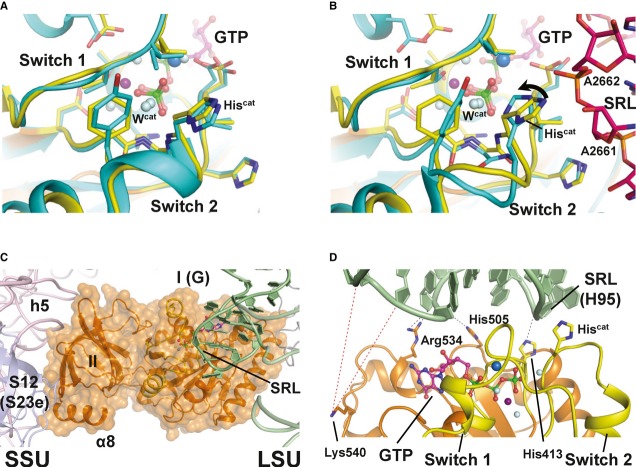
Figure 4. Structural model of eIF5B·GTP on the ribosome.
- Superposition of the catalytic centers of eIF5B·GTP (yellow) and free EF-Tu·GDPNP (cyan; PDB: 1EXM). Conserved residues are shown as sticks; GDPNP is omitted for clarity.
- Superposition of the catalytic centers of eIF5B·GTP (yellow) and ribosome-bound EF-Tu·GDPCP (PDB: 2XQD, 2XQE) with the sarcin-ricin loop (SRL) in pink. Structural alterations relative to free eIF5B·GTP and EF-Tu·GDPNP are limited to Hiscat of EF-Tu, which is reoriented (arrow) into its active position between A2662 and Wcat.
- Model of domains I and II of eIF5B (orange) on the ribosome, based on the superposition with EF-Tu·GDPCP. Similar to eIF5B·GDPCP in the cryo-EM model of the 80S IC (see Supplementary Fig S5B), the G domain is associated with the SRL of the large subunit (LSU; green), while domain II interacts with the body of the small subunit (SSU; light pink).
- Putative interactions between the G domain and the SRL/H95 (green). Direct interactions are indicated by black dashed lines; red dashed lines indicate the positions in H95 that are cleaved by Fe(II)-BABE introduced in the position of Lys540 (Unbehaun et al, 2007). His505 lies only 3.5 Å from H95, explaining why the H505Y mutation results in a reduced affinity for the ribosome and GTPase deficiency in eIF5B (Shin et al, 2002) (see also Supplementary Table S1). The conserved Arg534 likely contributes to the interactions with H95.