Figure 3. ORFscore discriminates translated from non-translated regions.
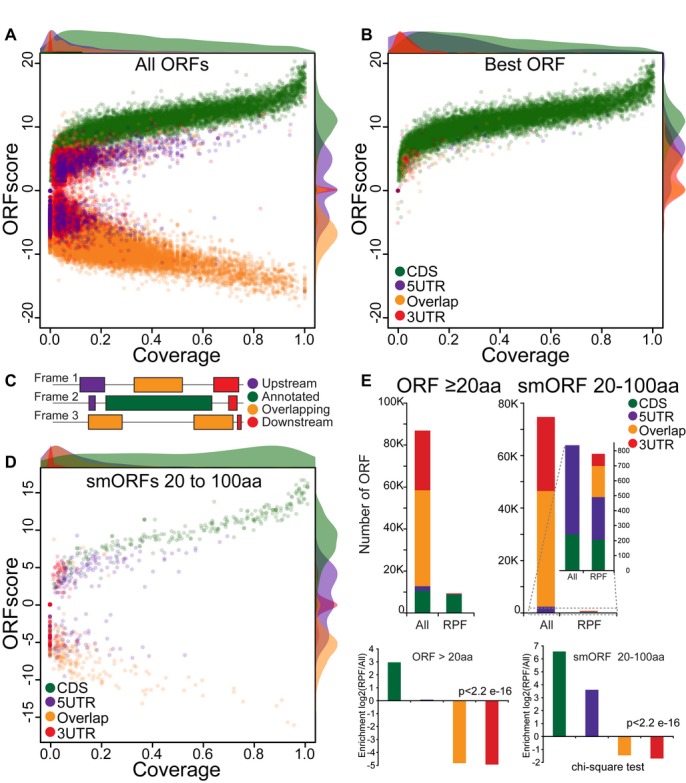
A–D Scatterplot of the ORFscore and coverage for all ORFs (A), the subset of ORFs with the highest ORFscore per transcript (B) and short (20–100 aa) annotated CDS (D). Relative density plots (scaled to the maximum value for each group) of the ORFscore and coverage are shown for each ORF type. Note the separation between annotated ORFs from the rest of the ORFs, even for short (20–100 aa) annotated CDSs. (C) Color code used to label different ORF types found in RefSeq protein-coding transcripts: annotated CDS (green), 5′UTR ORFs (purple), 3′UTR ORFs (red) and ORFs overlapping the annotated CDS (orange).
E Bar plots representing the number of ORFs identified on the basis of their ORFscore and coverage and defined as translated for each ORF type as in (C). Among all putative ORFs, the distribution of annotated ORFs was significantly different from the overall set (P = 2.2e-16, chi-squared test) with long and short CDS showing the highest fold-change enrichment in translated ORFs compared to other ORF types.
